evo-devo
@Xiaojie_Qiu
Followers
3,548
Following
1,539
Media
82
Statuses
895
Assistant Prof. @ Stanford BASE, Genetics & Computer Science (courtesy). Lead the predictive biology lab of ML & single cell/spatial genomics, focus on heart
Palo Alto, CA
Joined November 2015
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
SEVENTEEN
• 886372 Tweets
Manchester United
• 280737 Tweets
Manchester United
• 280737 Tweets
#MUFC
• 171434 Tweets
HAPPY 9TH ANNIVERSARY SVT
• 153080 Tweets
#SVT_Shinin9_Diamonds
• 146835 Tweets
Leclerc
• 138482 Tweets
Mainoo
• 112525 Tweets
Perez
• 81518 Tweets
Wembley
• 79064 Tweets
Garnacho
• 75031 Tweets
George Floyd
• 71629 Tweets
Martinez
• 70568 Tweets
Man City
• 68854 Tweets
Viva la Patria
• 53859 Tweets
Walker
• 53751 Tweets
Conference League
• 42907 Tweets
Rodri
• 42166 Tweets
#MCIMUN
• 37839 Tweets
Bruno Fernandes
• 35576 Tweets
Doku
• 33566 Tweets
Haaland
• 31555 Tweets
Onana
• 31527 Tweets
Varane
• 22039 Tweets
茨城県南部
• 21582 Tweets
Checo
• 19447 Tweets
マリノス
• 18809 Tweets
Amrabat
• 18667 Tweets
Licha
• 17440 Tweets
ユナイテッド
• 17090 Tweets
세븐틴 9주년
• 16736 Tweets
De Bruyne
• 15831 Tweets
السيتي
• 14920 Tweets
Gvardiol
• 13299 Tweets
#FACUP24
• 12774 Tweets
اليونايتد
• 10485 Tweets
Last Seen Profiles
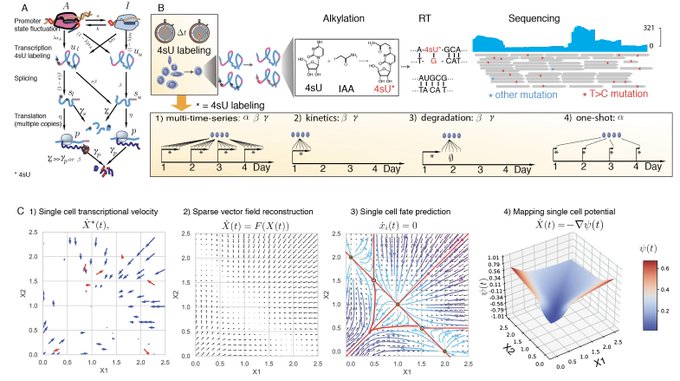
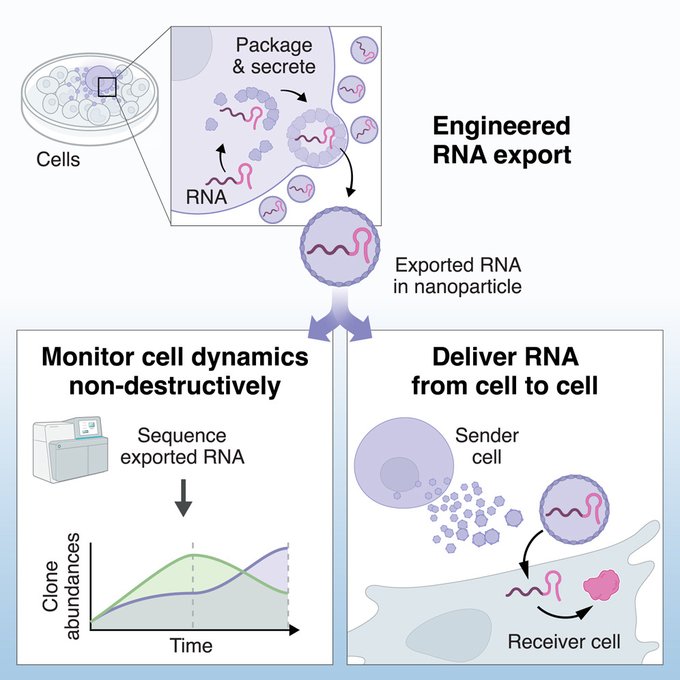
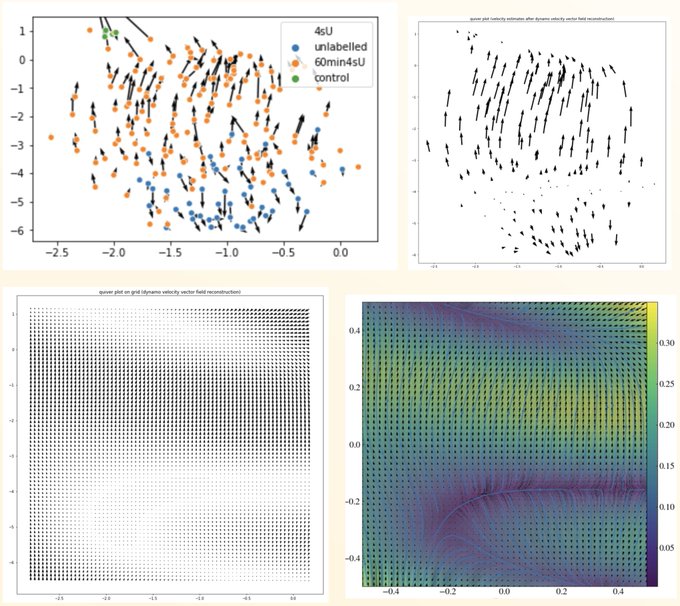
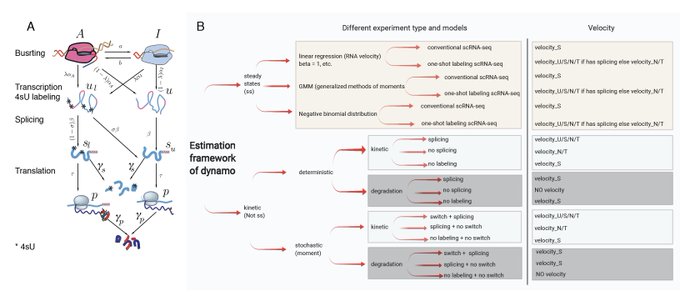
Wonder how to take RNA velocity to the next level with labeling based scRNA-seq? Check our new tutorials () on the time-resolved vectorfield analysis for cell cycle/intestine organoids of scEU-seq
@AlexandervanOu1
and neural activation of scNT-seq
@hao_wu_7
6
63
269
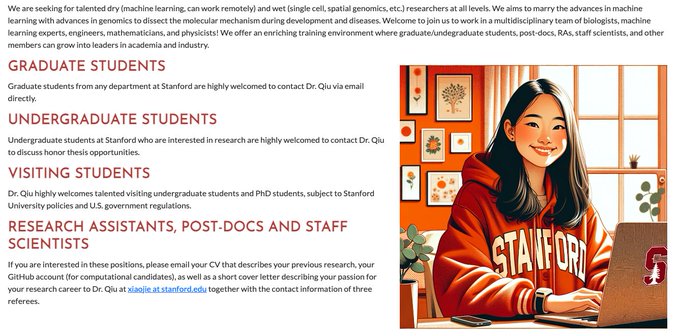
🔬My sc/spatial genomics lab (Launch at
#Stanford
on Dec. 16) is seeking experimental RAs & post-docs in genomics & dev bio. Come to work with me and a vibrant team of sys biologists, ML experts, etc. to push the boundaries of gene regulation of development & disease.
#HiringNow
1
71
216
Thrilled to share spateo-viewer: the “Google earth” App of spatial transcriptomics (ST) that allows interactive 3D alignment of ST, dynamic data exploration, etc. Kudos to Jiajun
@JiajunYao14
from Spateo development team on this beautiful work! More at:
2
41
140
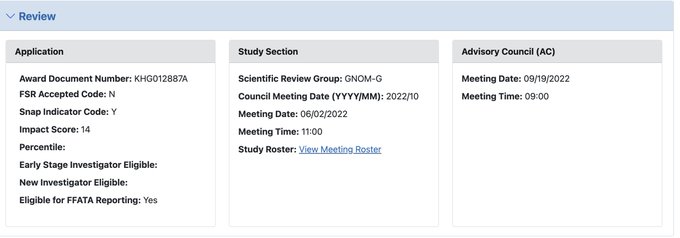
After a long time overdue, finally received the NOA for my K99 application (impact score of 14). Thanks for all supports from
@JswLab
, my collaborators of the grant
@hao_wu_7
@WangXiaoLab
and other mentors from my PhD/Post-doc,
@coletrapnell
@JShendure
@jhxing001
@bloodgenes
16
0
140
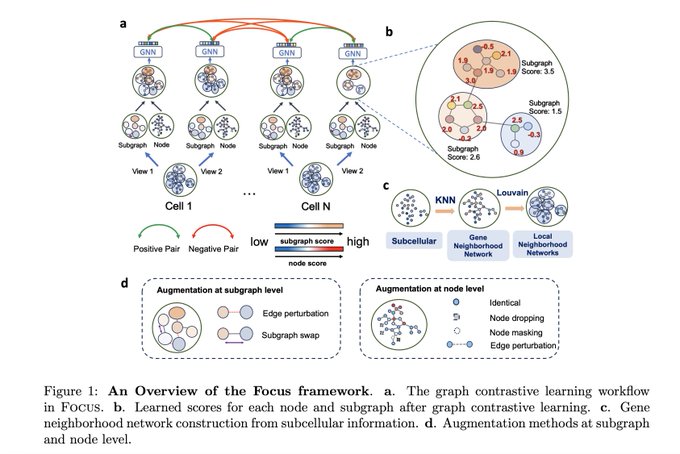
With the imaging-based spatial transcriptomics such as MERFISH, seqFISH, CosMx SMI, Xenium and others, have you ever wondered how we can leverage their subcellular spatial information? Check out our latest preprint on Focus by Qiaolin and Jiayuan
@JiayuanDing
, two talent
4
21
105
This is really overdue, but so glad that my last PhD work, Scribe (), was finally out. The finest work of my PhD career. Blessed to have Cole as the best advisor
@coletrapnell
. & very much thanks to
@sreeramkannan
and
@ARahimzamani
for the amazing colllabo
1
20
85
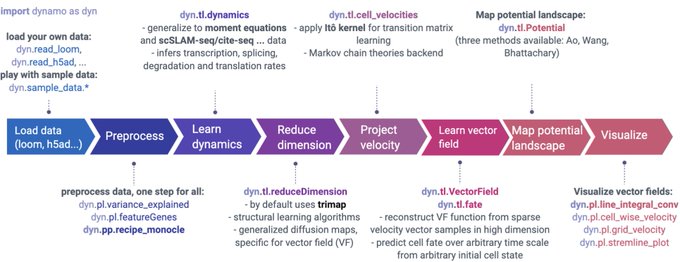
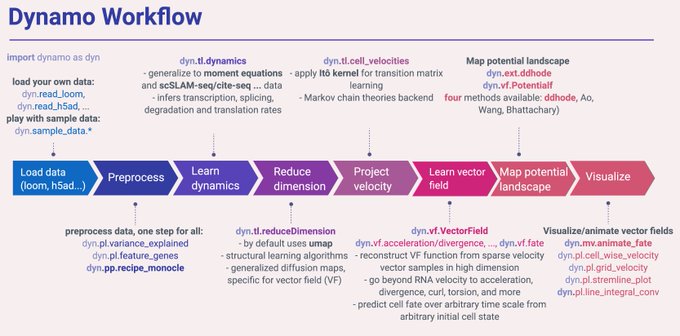
🚀 Excited to announce Dynamo v1.3 for advanced modeling of scRNA-seq and vector field learning & prediction! A big step towards our v2.0.0 goal by the end of 23'. Get it now on PyPi ()!
#RNAVelocity
. See fully refactored preprocessing module, more below
3
14
74
I am honored to be selected as Arc's inaugural Ignite Award recipient, among an amazing pool of colleagues. Really looking forward to joining the extended Arc family and interacting/collaborating with its core/innovation investigators & other recipients.
Congratulations to
@zhenanbao
,
@icclarker
, Nathanael Gray,
@WJGreenleaf
,
@BrianHie
,
@ishahjain
,
@AnnaMolofskyLab
,
@AlannaSchepartz
,
@yimmieg
, Katrin Andreasson,
@FaranakFattahi
,
@mfgrp
,
@MartinUCSF
, Calvin Kuo,
@MarsonLab
,
@brainevodevo
,
@Xiaojie_Qiu
, Julia Salzman,
1
1
15
6
3
68
Congrats, Felix! Mind-blowing work. This in principle solved the a key problem in single cell genomics that doesn't allow longitudinal observation of the same cell time. A much elegant solution than the LIVE-seq approach from nature recently!
1
9
48
Curious about how to generate an awesome animation like the one for the cool scNT-seq neural activation dataset () with dynamo? Now we updated our tutorial to show how to produce animation in a just a few lines of code here:
1/8 Want to capture temporal information about mRNA at single-cell levels? Check out our method, scNT-seq
@naturemethods
(). Developed by
@QiQiu8788
,
@PengHu_xqq
. Thanks to our collaborators:
@GovekKiya
@camara_pg
and
@Xiaojie_Qiu
.
6
92
277
2
10
35
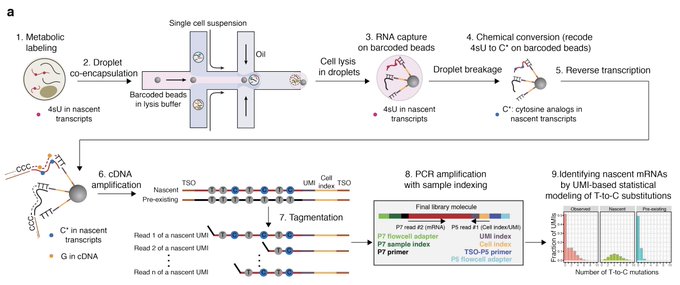
check out this amazing work from
@hao_wu_7
lab. nascent mRNA labelling in droplets! A must-read before the holiday season.
1
9
29
congrats! I am pleased to be one of reviewers for this simple but interesting approach to improve splicing RNA velocity estimation: it fits a NN for each gene with a cosine correlation loss which leads to gene-wise velocity nicely flows the curvature of the data in a phase plot
2
2
28
I am eagerly looking forward to working alongside my colleagues such as Jesse
@jengreitz
, Casey
@CaseyGifford
, Mark
@mascott85
, Marlene
@StanfordBASE
, Anshul
@anshulkundaje
, and Jure
@jure
, Bo
@PhysPlanaria
, and many others.
2
0
22
My brilliant collaborator Yan
@YanZhan92480937
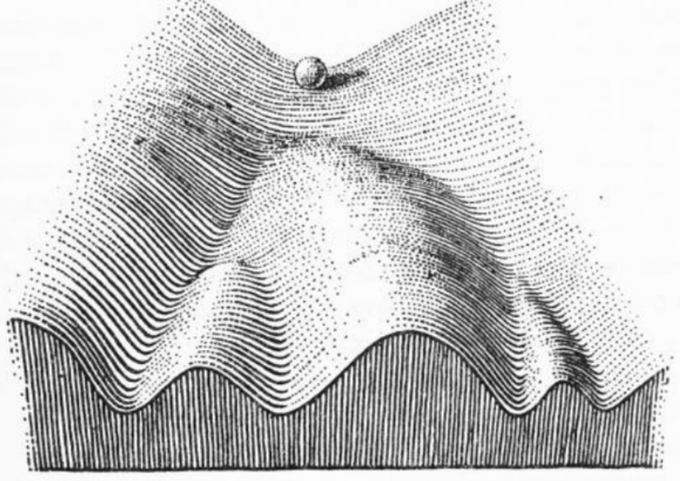
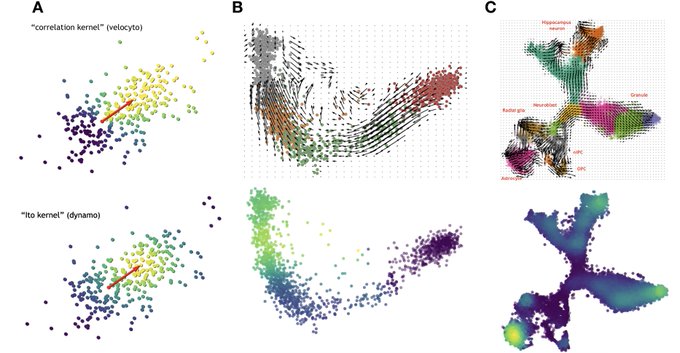
will talk about exciting updates of dynamo at . Dynamo now is powered by a principled approach for constructing a Markov transition matrix via "Itô kernel". Come to talk with him if u happen to be in the conf.
1
5
21
We will be a part of the Stanford BASE program
@StanfordBASE
, jointly with the Department of Genetics
@StanfordMed
, and affiliated with the Department of Computer Science
@StanfordEng
.
1
0
20
A special thanks to Sean
@SeanM_Wu
for inviting me to apply to BASE after noticing my work on spatial transcriptomics. Thank you so much!
1
0
19
My gratitude extends to all my previous/current funding supports such as the Jameel Clinic
@AIHealthMIT
, Impetus
@impetusgrants
, CZI
@czi
, K99
@NIH
, and others. Thank you all for believing in my work and vision.
3
0
17
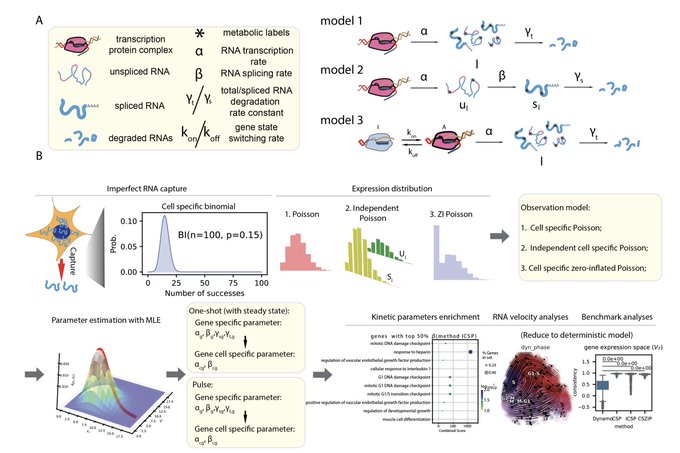
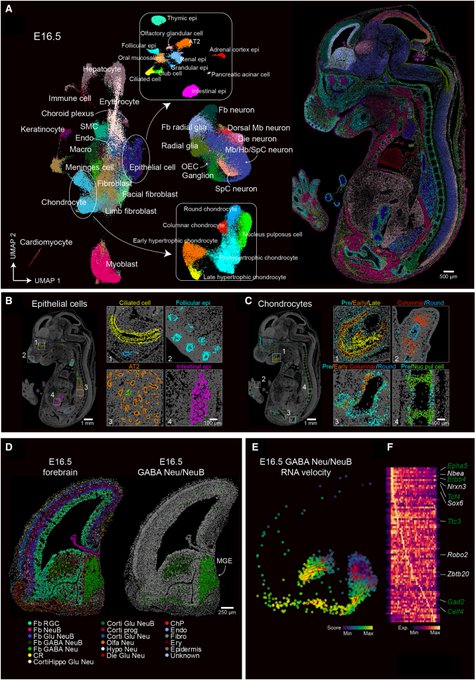
Amazing work from Wang lab at Broad on time-resolved, and spatially-resolved transcriptomics via TEMPOmap. See how they can quantify RNA kinetics over time in subcellular resolution for thousand of genes. Great honor to contribute to this amazing work.
Excited to share TEMPOmap, our new spatiotemporal transcriptomics method that integrates metabolic labeling with 3D in situ sequencing to simultaneously profile the age and location of individual RNA molecules led by
@JingyiRen
Haowen Zhou and
@huzengHZ
1
70
279
0
1
15
Thanks
@tsuname
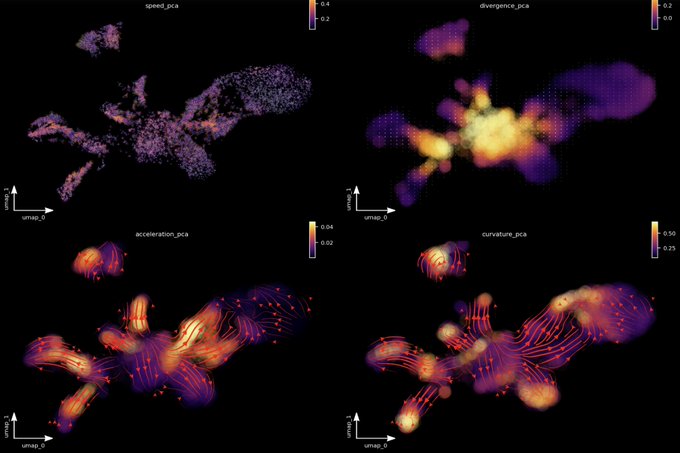
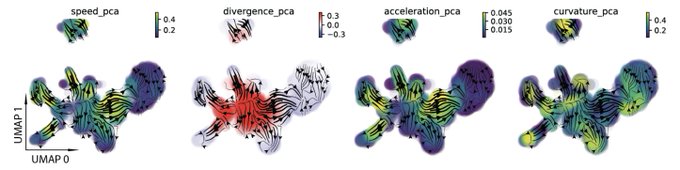
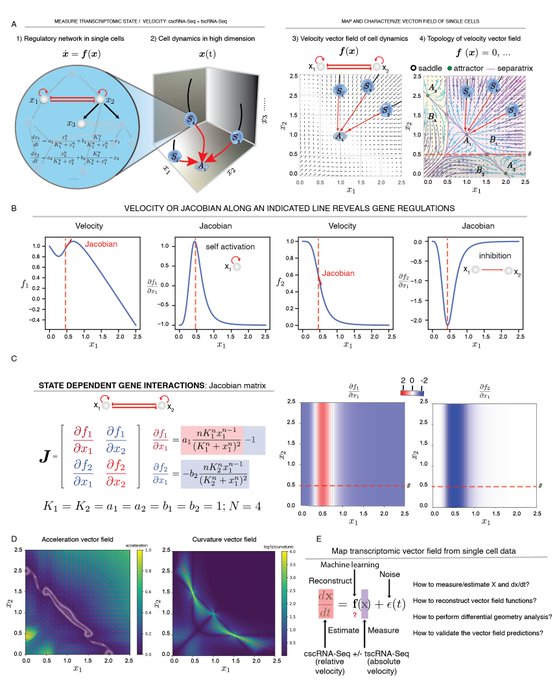
! Really unimaginable until very recently - now we can get those geometric quantities thanks to vector-field functions learnt from data with dynamo. Acceleration/curvature can be defined for each gene, you can use them just like expression or velocity matrices.
Nice connections between geometric concepts and cell biology in single-cell transcriptomic vector fields by
@Xiaojie_Qiu
. From
1
2
26
0
3
15
The list is long (chronological order) but includes Jay
@JShendure
(monocle2/3), Qi Mao (Monocle 2/3), Junyue
@junyue_cao
(monocle 3), Sreeram
@sreeramkannan
(Scribe, Dynode), Arman (Scribe, Dynode), Jianhua
@jhxing001
(Dynamo), Yan
@YanZhan92480937
(Dynamo), Dian (dynamo)
@dian
1
0
14
Hao
@hao_wu_7
(scNT-seq), Qi
@QiQiu8788
(scNT-seq), Joseph
@lioscro
(Dynast, Spateo), Jorge
@jmartinrufino
(Dynamo), Chen
@chenweng1991
(Dynamo), Daniel
@DanielYZhu
(Spateo), and many others. (Forgive me if I may miss anyone here. )
2
0
14
I would be remiss not to mention the invaluable support, guidance, and mentorship I received from my post-doc advisor
@JswLab
my PhD advisor
@coletrapnell
, and a host of incredible collaborators over the years.
1
1
13
Really enjoyed the collaboration with the amazing
@hao_wu_7
@QiQiu8788
@PengHu_xqq
! Also very proud to show dynamo can take full advantage of scNT-seq to recover "time-resolved RNA velocity" of neural activation while conventional scRNA-seq cannot!
1/8 Want to capture temporal information about mRNA at single-cell levels? Check out our method, scNT-seq
@naturemethods
(). Developed by
@QiQiu8788
,
@PengHu_xqq
. Thanks to our collaborators:
@GovekKiya
@camara_pg
and
@Xiaojie_Qiu
.
6
92
277
3
0
12
congrats
@WangXiaoLab
@JingyiRen
@hzhou99
. Honored to contribute to this amazing work to track the RNA production, nuclear exportation, trafficking and degradation at subcellular resolution with the TEMPOmap.
Congrats to
@JingyiRen
,
@hzhou99
, and the great team! With TEMPOmap, we can snapshot RNA life cycle from birth to death for thousands of genes with single-cell and subcellular resolutions, revealing that RNA kinetics, fast and slow, serves gene functions.
9
17
154
0
0
11
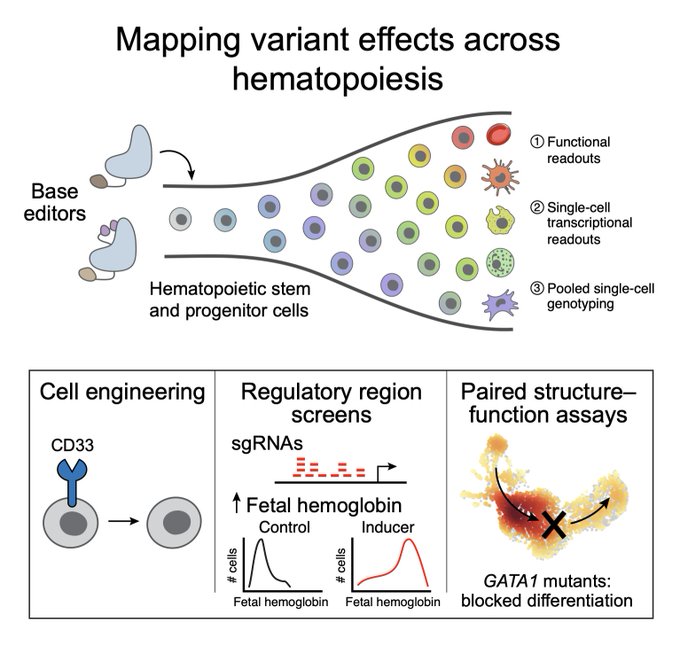
Super cool work from
@jmartinrufino
in Cell to massive parallel creating genetic variants across genes and coupled w/ scRNA-seq to reveal downstream effects w/ many clinical potential. Delighted to contributed to the downstream velocity analyses w/ dynamo
Thrilled to share my PhD work in
@CellCellPress
! Massively parallel base editing to map variant effects in human hematopoiesis. We developed screens on blood stem cells to understand disease and develop treatments at single-nucleotide resolution.1/n
15
99
434
0
1
10
Also greatly thanks to
@chenweng1991
@jmartinrufino
from labs
@bloodgenes
@eric_lander
who helped generate the human hematopoiesis scNT-seq dataset during the revision processes which fundamentally reshape the fate of the work.
1
0
6
@slinnarsson
@NimwegenLab
the RNA-velocity paper is definitively one of the most exciting paper I ever read in the single cell genomics field. It is based on a simple model but the results are surprising great. I am shocked that it is rejected.. best wishes for the appeal
0
2
7
Of course, I would greatly thank the pioneers of RNA velocity and all other recent exciting new developments
@GioeleLaManno
@slinnarsson
@KharchenkoLab
@fabian_theis
@dana_peer
@VolkerBergen
! (We are deeply regretful if we missed anyone here. )
1
0
6
Thanks for funding this research!
Jameel Clinic PI Jonathan Weissman (
@JswLab
), postdoc
@Xiaojie_Qiu
, and collaborators at the University of Pittsburgh School of Medicine are working on a ML framework called "dynamo" that will help predict cell fates and genetic perturbations.
Read more:
0
3
4
0
0
6
Welcome to Jsw lab for a new exciting chapter in your career!
It feels bittersweet to finish my last week in
@davidasinclair
lab. I was so lucky to find this dream match in the 4th rotation and start an incredible 6y journey with some low lows and high highs🍾. Now onto the next chapter with
@JswLab
to keep learning and growing💪. Excited!
16
4
301
0
0
6
My great thanks also go to
@shayanhoss
from
@spyros_darmanis
lab who contributed tremendously to help generate the clonally-traced scSLAM-seq/NASC-seq dataset used for the initial submission.
2
0
6
The Scribe manuscript is finally out on biorxiv after an epic delay! very proud of it, one of my best in my PhD. We show the importance of dynamics time coupling for causality detection. Hope this message will lead to a new era of research. Great collaboration with
@sreeramkannan
0
0
5
@chapin_jen
@fengsylmar
真他妈丢人,5000不叫历史?你美国爸爸就300年。今天偶尔看到在twitter上有这么弱智的人在骂中国,中国禁止屏蔽这些弱智的言语闷声发大财真是正确的。
0
0
4
@SteveO_in_AZ
@PostBio
@SpokespersonCHN
@RichardGrenell
I really hope you are among the millions of death.
1
0
5
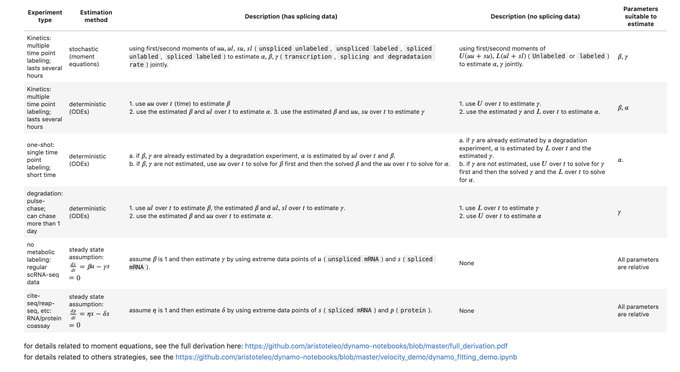
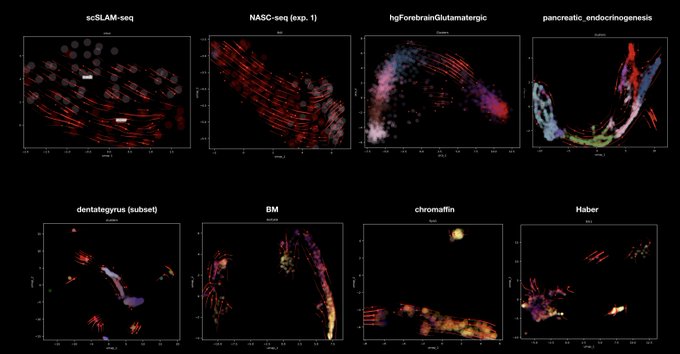
⏩ For the next release Dynamo v1.4, we'll bring new methods for metabolic labeling enabled, time-resolved
#scRNAseq
(e.g. scSLAM-seq, NASC-seq, scEU-seq, sci-fate, scNT-seq) + more enhancements on vector field learning. Stay tuned!
#bioinformatics
#RNAseq
1
2
5
Thanks
@tsuname
for speaking very highly of the conceptual innovation of our work! Bridging small-scale systems-biology/physics type of thinking with high-dimensional genomics using ML is my long-time dream. We are still pushing the limits of our continuous vector field approach
0
0
5
Spateo is our latest efforts to build Aristotle ecosystem, a novel full-stack computational ecosystem that provides advanced spatiotemporal modeling of single cell and spatial genomics datasets. This work is supported by
@cziscience
() and
@impetusgrants
1
1
5
Congrats on this amazing work on revealing the single-cell clinical COVID19 neutrophil response and dexamethasone treatment kinetics
@sarthak_sin
@jeffbiernaskie
. Nice application of the analytical vector field approach from dynamo ()
Tour de force from
@sarthak_s
in
@jeffbiernaskie
Lab and
@NicoleLRosin
mapping the
#COVID19
clinical neutrophil response and the effects of Dexamethasone
Great application of Dynamo by
@Xiaojie_Qiu
Potentially of interest
@MattMeiz
@KaminskiMed
@GreaneyAllison
@DelaCruzYaleMed
3
1
6
0
1
5
Come to work in the Lander lab at the Broad. An amazing opportunity for recent undergraduate graduates who want to work on some exciting technology development projects before applying PhD. Contact
@jmartinrufino
for more details
0
0
5
I am extremely thankful for
@JswLab
incredible mentoring and support over the past few years. This work is not possible without
@YanZhan92480937
(co-first author) from
@jhxing001
lab who contributed enormously to the theoretical undertaking of this work.
1
0
4
Congrats Liangcai
@GuLiangcai
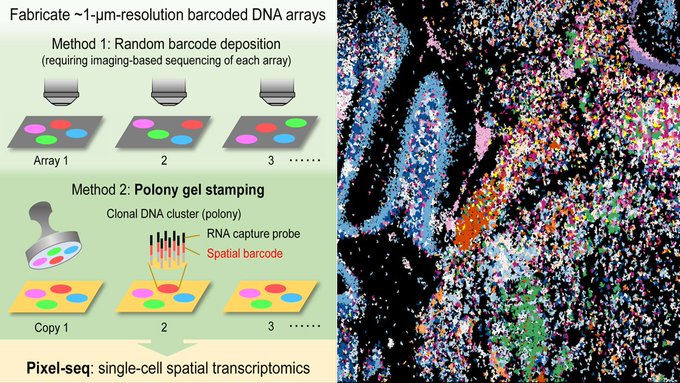
, Xiaonan, Li and Runze and all others for this amazing work! PIXEL-seq with polony gel stamping that dramatically reduce time andcost will be another game changes in single-cell or subcellular resolution spatial transcriptomics!
Our polony stamping & Pixel-seq paper is online! Congrats to our great team! Polony gels were designed for but will go beyond spatial omics.
@UWBiochemistry
@uwgenome
@NAPECenter
@UWPharmacology
@UWAnesthPainMed
@SpatialOmics
#ChronicPain
#SingleCell
1
19
51
0
2
5
@benoitbruneau
@BGI_Research
@MLongqi
currently stereo-seq can be accessed via research collaboration with BGI (you can directly email Longqi: liulongqi
@genomics
.cn). There are already kits for it and plans for commercializing it soon. it should be able to sequence the library with illumina too but
2
0
4
So implied in the logo, dynamo utlizes scRNA-seq, especially metabolic labeling enables ones, to learn the functions of vector fields to make non-trivial mechanistic and predictive insights: . Treat thanks to
@dummyIndex
and the team for this elegant design
2
1
4
As always, we welcome any comments & suggestions on our work. As I am starting my lab at Stanford & actively looking for talented research assistant/post-doc, etc., if you are interested in these kind of research, please email me at xqiu
@mit
.edu. Excited to working with you!
0
0
4