Jonathan Weissman's Lab
@JswLab
Followers
11K
Following
338
Media
12
Statuses
147
Jonathan S. Weissman's lab at the @WhiteheadInst/@MIT. Account run by lab members.
Cambridge, MA
Joined October 2019
Excited to share new work led by @AniaPuszynska on how lysosomes are altered during aging.
Excited to share our work with @DMSabatini & @JswLab. How do lysosomes change with age? We present a metabolic atlas of lysosomal aging, and reveal a lysosomal “aging clock” of metabolites linked to lysosomal storage disorders. Grateful to all co-authors! https://t.co/HyP6zq3Ccq
0
1
19
Exciting work from the lab, on the mechanism of OSK rejuvenation, led by @Y_Ryan_Lu with lab co-authors James Cameron, @MohamedBrolosy, @AniaPuszynska, @Xiaojie_Qiu.
1/ Can we find more precise and even safer ways to rejuvenate vision? As a step towards this goal, I am excited to share this preprint of my 1st postdoc work from @JswLab with amazing collaborators, esp. @Bruce_Ksander! Using functional genomics, we discovered GSTA4 as a key OSK
1
6
28
Exciting work from the lab led by @JingchuanLuo with coauthors @KhandwalaStuti @LevineZebulon and JJ Hu
I'm thrilled to share our work from @JswLab ( https://t.co/CMzQznSXPN). We developed LOCL-TL, an optogenetic approach for monitoring localized translation in mammalian cells. LOCL-TL revealed two mechanistically distinct strategies for mitochondrially localized translation.
1
5
40
500K builders use YouWare to build ideas for their side hustles and client projects. Join the movement + follow for daily inspiration.
0
1
0
Excited to announce that this work is now live as a first release article @ScienceMagazine So grateful for this fantatic team and to see this work in wild!
science.org
Charting the spatiotemporal dynamics of cell fate determination in development and disease is a long-standing objective in biology. Here, we present the design, development, and extensive validation...
1/14 On behalf of the amazing team in @JswLab, we’re excited to share PEtracer ( https://t.co/QTyBahCCLS) a prime editing-based evolving lineage recorder compatible with both scRNA-seq and high-resolution imaging readouts in intact tissue. By applying PEtracer in a syngeneic mouse
6
18
91
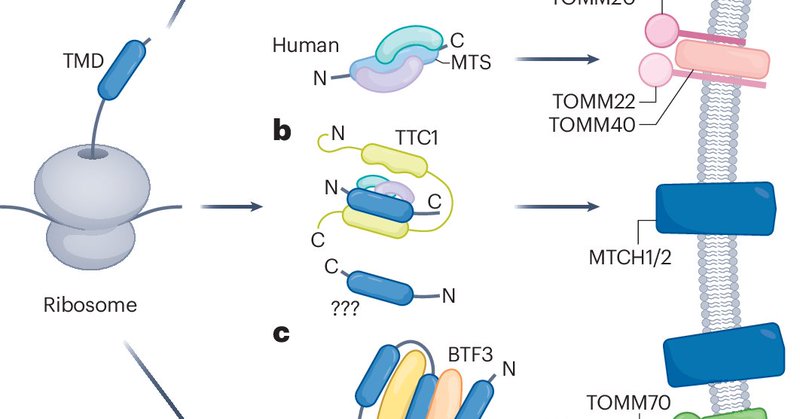
Really enjoyed writing this review on the recently expanded landscape of human outer mitochondrial membrane protein biogenesis, its evolution across eukaryotes, and comparisons to the ER biogenesis systems! @JswLab
https://t.co/TCn8OmohwT
nature.com
Nature Cell Biology - This Review discusses the mechanisms underlying the insertion of α-helical proteins into the mitochondrial outer membrane and highlights the role of convergent evolution...
3
10
72
Hear it straight from our amazing first author @_annhuang to dive into the X-Atlas/Orion dataset and the FiCS Perturb-seq Platform. Excited to share our work from @Xaira_Thera. ( https://t.co/NaPLtqX6MA)
1/12 Excited to share our team's latest work and the first @xaira_thera preprint! Here, we introduce FiCS Perturb-seq, an industrialized platform for generating scaleable, high-quality perturbation data. 📄 Read the preprint:
0
2
8
1/14 On behalf of the amazing team in @JswLab, we’re excited to share PEtracer ( https://t.co/QTyBahCCLS) a prime editing-based evolving lineage recorder compatible with both scRNA-seq and high-resolution imaging readouts in intact tissue. By applying PEtracer in a syngeneic mouse
1
3
9
1/14 On behalf of the amazing team in @JswLab, we’re excited to share PEtracer ( https://t.co/QTyBahCCLS) a prime editing-based evolving lineage recorder compatible with both scRNA-seq and high-resolution imaging readouts in intact tissue. By applying PEtracer in a syngeneic mouse
biorxiv.org
Charting the spatiotemporal dynamics of cell fate determination in development and disease is a long-standing objective in biology. Here we present the design, development, and extensive validation...
3
57
195
Congrats @NadigAjay and @Luke0connor!
Congrats @NadigAjay on TRADE out now in @NatureGenet: https://t.co/eHwDZEIqNd These statistical metrics enable more meaningful comparisons in Perturb-seq atlases. Also, now find the HepG2 and Jurkat Perturb-seq datasets on GEO GSE264667!
0
0
6
Congratulations to James Numez and collaborators for their RENDER platform for delivering epigenetic editors using engineered VLPs. RENDER broadly advances our ability to deliver epigenome editors into human cells and will likely find ciritcal uses both in fundamental and
biorxiv.org
Programmable epigenome editors modify gene expression in mammalian cells by altering the local chromatin environment at target loci without inducing DNA breaks. However, the large size of CRISPR-ba...
1
2
35
Delighted to share this preprint from Prof. Rong Fan Lab @RongFan8 and Prof. Sidi Chen lab @sidichen on Spatially Resolved in vivo CRISPR Screen Sequencing via Perturb-DBiT. Congatulations, Alev, Xiaolong, Feifei, Paul, Sidi and Rong!
Thrilled to share Perturb-DBiT — spatial unbiased in vivo perturb-seq with genome-scale CRISPR libraries! Hope you enjoy reading it over the Thanksgiving week 🥰 Kudos to @AlevBaysoy, Xiaolong, Feifei, and Paul. & collaboration with @sidichen, Hongbo Chi. https://t.co/IcUPAG5k8P
1
3
22
Excited that the Zhuang Lab is now on X!
Check out our preprint on Perturb-multi ( https://t.co/lFeRYA0lco), a multi-year collaborative effort that enabled multi-modal in vivo genotype-phenotype mapping by pooled genetic perturbations and high-dimensionality phenotype readout by imaging and sequencing.
0
1
9
Our wonderful collaborators on the Perturb-Multi work ( https://t.co/P64ppY3x13) are now here and on the other app, at @ZhuangLab
1
5
46
In collaboration with Reuben Saunders, @JswLab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! https://t.co/iJ8hi3ddz4
biorxiv.org
Organ function requires coordinated activities of thousands of genes in distinct, spatially organized cell types. Understanding the basis of emergent tissue function requires approaches to dissect...
2
116
339
Very exciting new work on multimodal, in vivo perturb-seq!
In collaboration with Reuben Saunders, @JswLab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! https://t.co/iJ8hi3ddz4
0
8
132
We hope that our fixed-cell methods will enable a new era of large-scale in vivo Perturb-seq!
0
0
5
The data quality from fixed-cell Perturb-seq is at least as high as that we have seen with RT-based scRNA-seq, and the economics are significantly improved due to the droplet overloading enabled by the probe barcodes from @10xGenomics.
1
0
7
Fixed-cell Perturb-seq is much easier and less stressful than live cell Perturb-seq—both for the user and for the cells! We love working with samples that can sit in the fridge for days with minimal degradation.
1
0
4
We then hybridize the fixed cells to custom split probes that directly bind to our sgRNAs, alongside the transcriptome-wide mRNA libraries from @10xGenomics.
1
0
4
We harvest mosaic mouse tissues by perfusion with formaldehyde, which immediately locks transcriptional phenotypes in place and protects RNA. We then dissociate the fixed tissue and use FACS to select perturbed cells.
1
0
3