Chen Weng
@chenweng1991
Followers
433
Following
389
Media
20
Statuses
114
Postdoc in Sankaran lab and Weissman lab @bloodgenes & @JswLab || Single-cell genomics/Cell-fate commitment/Gene regulation/
Cambridge, MA
Joined February 2012
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
Marcellus Williams
• 349170 Tweets
Missouri
• 210839 Tweets
#FreetheCitizens
• 121754 Tweets
Vinicius
• 119306 Tweets
OQTF
• 78261 Tweets
Mike Parson
• 68008 Tweets
Alavés
• 64082 Tweets
#StateofPlay
• 48599 Tweets
Endrick
• 48089 Tweets
Horizon
• 47523 Tweets
Ghost of Yotei
• 45635 Tweets
発売日決定
• 41943 Tweets
#GHLímite3
• 40526 Tweets
Manchin
• 39750 Tweets
Parkinson
• 32012 Tweets
ラヴィット
• 30113 Tweets
Lunar
• 29293 Tweets
Quito
• 27630 Tweets
Vallejo
• 25825 Tweets
アルシュベルド
• 19423 Tweets
メインモンスター
• 19375 Tweets
EFCC
• 17276 Tweets
ワイルズ
• 14061 Tweets
アイドル19歳
• 12485 Tweets
Last Seen Profiles
Pinned Tweet
Very delighted to share our work Deciphering cell states and genealogies of human hematopoiesis online today in
@Nature
(Accelerated Preview version). It is a really great collaboration of Weissman lab
@JswLab
and Sankaran lab
@bloodgenes
🧵(1/n)
11
72
285
Delighted to share our work studying the molecular control for the directed β cell differentiation process
@NatMetabolism
Great collaboration with Jiajia Xi
@FulaiJin
lab and Yan Li lab
@CWRUSOM
.
3
7
50
Delighted to share the last piece of my graduate school work that dissects disease-associated single-cell heterogeneity and continuum through multiomics
@NatureComms
.
#SingleCell
#Multiomics
#diabetes
4
10
45
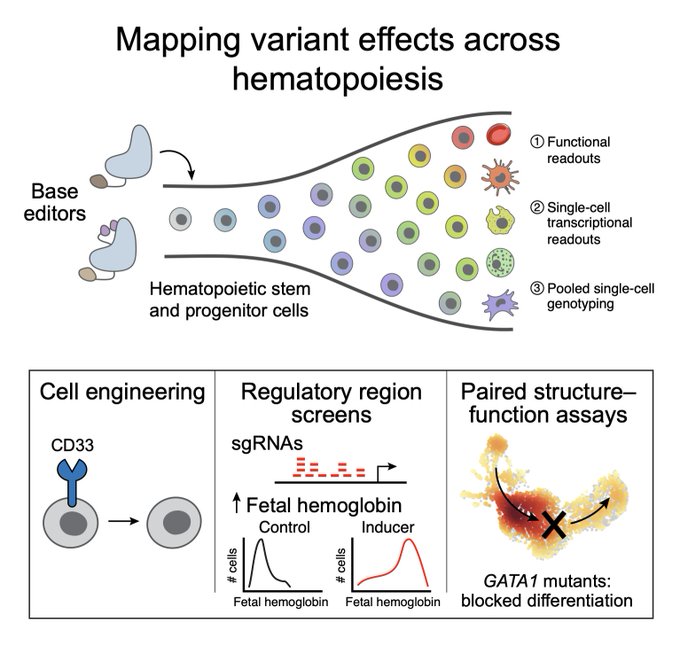
Massively parallel base editing to map variant effects in human hematopoiesis. Incredibly powerful to study variant-to-function at scale with single-nucleotide resolution in primary human cells. Amazing work!
@jmartinrufino
@bloodgenes
Thrilled to share my PhD work in
@CellCellPress
! Massively parallel base editing to map variant effects in human hematopoiesis. We developed screens on blood stem cells to understand disease and develop treatments at single-nucleotide resolution.1/n
15
99
440
0
1
20
I'm truly grateful for the rigorous, supportive, and constructive environment around me. Special thanks to my co-mentors
@bloodgenes
@JswLab
for their guidance, and heartfelt appreciation to my colleagues and family for their invaluable support!
1
0
15
@alexa_schnell
@KatieEYost
@LukeKoblan
@jmartinrufino
, many thanks to reviewers for their helpful suggestions, and also thanks for the very inspiring pioneering work on mitochondrial lineage tracing from
@LeifLudwig
@CalebLareau
, and colleagues. (end)
3
0
9
Excited to be presenting our work reconstructing single cell developmental tree for human pancreatic beta cell differentiation at
#ashg19
9am in level 3, ballroom C!
1
1
6
By way of background, we had described the single-cell Regulatory multi-omics with Deep Mitochondrial mutation profiling (ReDeeM) approach in a
@Nature
paper earlier this year: (2/n)
1
0
7
Our work on disease associated single-cell trajectory in pancreatic beta cell combined with genome-wide CRISPR screen is out
@CellReports
! Great collaboration with Zhou. Check our RePACT for disease trajectory analysis
0
3
6
I’m immensely grateful to my amazing joint mentors
@JswLab
@bloodgenes
and all collaborators and colleagues, in particular
@vangalenlab
Sahand Hormoz lab,
@fulong_yu
@dianyang1117
@michaelpoeschla
@la_liggett
@MattG_Jones
@Xiaojie_Qiu
@LWahlster
Alexis Caulier
@jeff_hussmann
…
1
0
5
Open full-text link to this work is available here:
Delighted to share our work studying the molecular control for the directed β cell differentiation process
@NatMetabolism
Great collaboration with Jiajia Xi
@FulaiJin
lab and Yan Li lab
@CWRUSOM
.
3
7
50
0
0
4
Yang lab hiring
@Columbia
State-of-art tracking and recording technologies in understanding tumor evolution and cancer plasticity
Excited to announce the Yang lab officially opens at
@Columbia
University. We are interested in developing innovative single cell recording technologies to study tumor evolution and cancer plasticity. We are hiring motivated postdocs & students, please RT
34
84
414
0
0
5
Impressive work to demonstrate purifying selections of mitochondrial pathogenic mutations in cell-state specific manner! Congrats
@CalebLareau
@LeifLudwig
and colleagues!
Out today in
@NatureGenet
, our work utilizing single-cell multi-omics to define specific immune cell subsets that experience purifying selection against pathogenic mitochondrial DNA. Check out a thread about this work below: 1/22
14
82
325
0
0
4
So excited for you, my friend! Xiaojie Qiu is starting his own lab at Stanford Dec 16th! He is working on really cool stuff in spatial-temporal modeling of single cell genomics in development. His lab is recruiting! Please reach out to him if you are interested!
0
0
4
@Li_Li_666
@YanHu41082612
@ShouwenW
@FD_Camargo
@BCHStemCell
@HSCRB
@harvardstemcell
Congratulations, Lili and colleagues!
1
0
1
Deeply grateful to my mentors
@FulaiJin
and
@yan_yan_li
for their guidance. A huge shoutout to co-first authors
@axg826
and
@shanshanzhang
for their relentless dedication, and a big thank you to all our colleagues for their invaluable contributions.
1
0
2
@jmartinrufino
@bloodgenes
@frdelpino
@DFBC_PedCare
@BBS_Harvard
@broadinstitute
Huge congrats, Jorge!!
0
0
2
Congratulations, Julia! Incredible resource!
Thrilled to share my primary PhD work: a Transcription Factor Atlas for understanding gene regulation and cell engineering
@CellCellPress
. We created a comprehensive TF ORF library and applied it to profile resulting expression changes. A thread 1/X
27
320
1K
1
0
2
Interesting idea(attempt) to study a phenotype thcaused by multiple genes in complex relationships
PRISM, a new
#CRISPR
#screening
technology, enables unbiased perturbations of
#transcriptionalnetworks
. PRISM was used to reveal genes that protect models of
#Parkinson
’s disease from
#alphasynuclein
-induced toxicity.
@MolecularCell
-->
0
64
103
0
0
1
@JiaruiMi
Thanks! That's a great question! S4C is the population transcriptionally most closely bridge to both endo/non-endocrine. And S4C actually shows PDX1+/SOX9+/GATA4-. So I think S4C is very likely the "bipotent trunk". But we'd need lineage/clonal info to make sure of that.
0
0
1
@carninci
Very interesting! Wonder the fraction of rRNA using the random primer? Any tricks needed to avoid the rRNA to saturate the library? Thanks!
0
0
0
@colonelragnar
@Nature
@JswLab
@bloodgenes
Thank you
@colonelragnar
!!! we are excited about both normal tissue homeostasis from adult stem cells and disease progression such as cancer evolution.
0
0
1
Interesting story
@eLife
on gain of enhancers by MMR-mutations in colon cancer. Congratulations, my friend Stevephen Hung!!
Check out our lab's latest paper
@eLife
on MMR-mutations as enhancer activators in colon cancer. Couldn't be more pleased with their review process. Special congrats to lead author and MD-PhD student Stevephen Hung. As always, feedback is welcome.
3
12
47
0
0
1