Jimmie Ye
@yimmieg
Followers
3K
Following
45
Media
57
Statuses
278
Professor @ UCSF, Genomicist, Geneticist, Adopted Immunologist, #GirlDad, Immigrant
San Francisco, CA
Joined June 2012
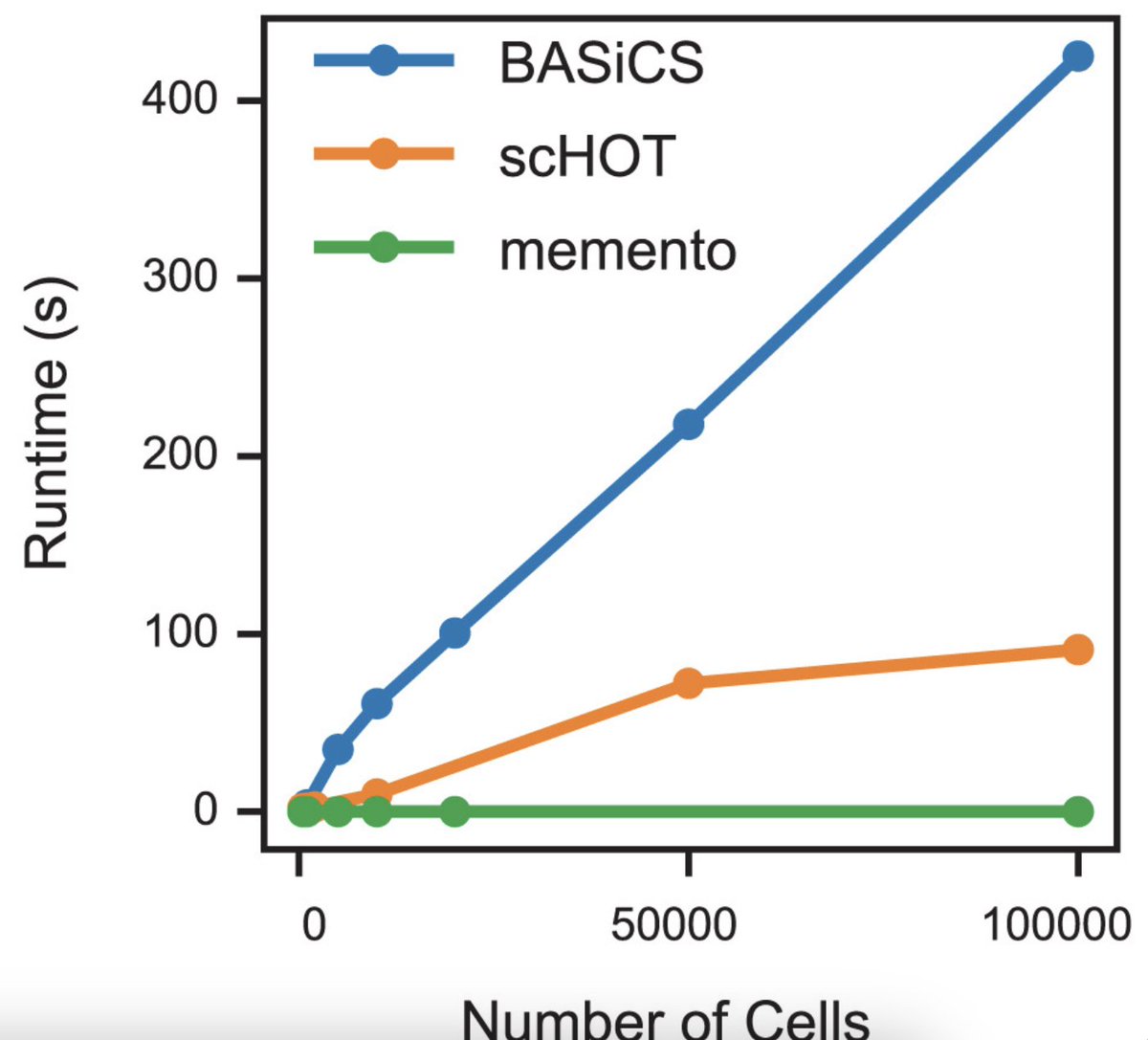
12/🧵One more thing: Memento’s shuffling strategy can be precomputed, allowing for deployment on massive datasets like @cziscience's 50 million cell atlas. We showcased its utility with the @cellxgene Discover API, enabling near-instant comparisons across diverse cell groups.
1
0
1
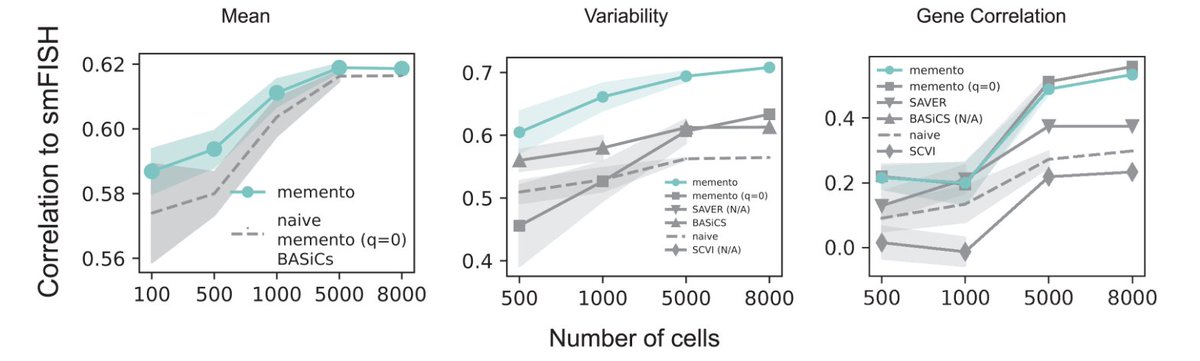
7/🧵In Memento, we leverage method-of-moments (MoM) estimators for the mean, residual variance, and gene correlation from scRNA-seq data. Memento’s estimates are better correlated with smFISH data generated by @arjunrajlab ( than imputation-based methods.
1
0
1
5/🧵We were inspired by @KleinLabHMS's InDrop paper that used the hypergeometric to model noise in scRNA-seq ( S19) and Zhang et al. that used a Poisson approximation (.
1
0
0
3/🧵@MattG_Jones initially experimented with a “minipseudobulk” approach to improve variance estimates. Our strategy was very similar to the MetaCell framework from Baran et al. However, minipseudobulk was a heuristic, not a solution.
1
1
2
RT @J_E_Mitchel: It's great to see my first paper of the PhD published in @NatureBiotech! This describes our scRNA….
0
48
0
RT @GladstoneInst: 🚨 NEW PUBLICATION 🚨 "Systematic decoding of cis gene regulation defines context-dependent control of the multi-gene cost….
0
6
0