Fei Chen Lab
@insitubiology
Followers
2,850
Following
107
Media
9
Statuses
122
Fei Chen's lab at @BroadInstitute and @HSCRB | Spatial and temporal genomics | Tweets by lab members | Where is Fei?
Cambridge, MA
Joined December 2021
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
Ireland
• 365634 Tweets
#WeAreSeriesEP8
• 352151 Tweets
Norway
• 315117 Tweets
Spain
• 272714 Tweets
#MissUniversePhilippines2024
• 155834 Tweets
Apple Music
• 134961 Tweets
PondPhuwin WeAre EP8
• 130430 Tweets
Blonde
• 89099 Tweets
LOST TEASER
• 88714 Tweets
KATY PERRY
• 67086 Tweets
Irlanda
• 54961 Tweets
Noruega
• 49693 Tweets
PREPARE FOR MLMU
• 47265 Tweets
#RCBvsRR
• 45866 Tweets
Rishi Sunak
• 39710 Tweets
KING KONG MV TEASER
• 38163 Tweets
Thriller
• 37402 Tweets
Atalanta
• 36760 Tweets
Lauryn Hill
• 32936 Tweets
مورينهو
• 31154 Tweets
Maxwell
• 24297 Tweets
Frank Ocean
• 24034 Tweets
TPAB
• 15569 Tweets
ガッキー
• 14937 Tweets
$BULL
• 14525 Tweets
Sokak Köpekleri Toplatılsın
• 12115 Tweets
#GeneralElection
• 12051 Tweets
GKMC
• 11748 Tweets
Ashwin
• 10345 Tweets
Last Seen Profiles
Buy your grad students a 3D printer, they said
It will be useful for science, they said
wondering what's the next big thing after spatial transcriptomics?
3D printed tube holders
@insitubiology
23
62
1K
14
29
716
Started at the bottom (
@broadinstitute
10th floor), now we're here (Broad 11th floor) 🚀
Celebrating the opening of our new lab space with a DIY ribbon cutting ceremony
9
6
403
Excited to share two new papers, published today in
@Nature
!
Check them out here:
(1) Slide-tags, a new method to unite single-cell and spatial profiling:
4
99
370
Our spatial transcriptomics DE method, C-SIDE, w/
@rafalab
+ Dylan Cable is now out in Nature Methods! Accounts for spatial mixtures of cell types to find cell type-specific DE as a function of e.g. pathology or other cell types.
4
80
284
Thanks to
@naturemethods
for writing this fantastic Research Briefing on C-SIDE, our new differential expression method for spatial transcriptomics!
0
24
151
Put this on your RADARS: our latest work on Reprogrammable ADAR Sensors for cell type targeting
A team effort by Chen lab members Jeremy Koob,
@dawnchenx
, & Yifan Zhang in collaboration with
@idmjky
,
@RohanKrajeski
,
@jgooten
,
@omarabudayyeh
, & AbuGoot lab
1
28
146
RADARS out in
@NBT
! Fantastic team effort with
@jgooten
@omarabudayyeh
and our lab:
@idmjky
,
@jgkoob
,
@dawnchenx
,
@RohanKrajeski
, and Yifan Zhang with
@VerenaVolf
,
@91_naynay
, Samantha R. Sgrizzi, and
@Lukas
Villiger
. Check out the manuscript here:
1
15
115
Interested in spatial transcriptomics analyses but don't know where to get started?
Check out this very practical walk-through for analyzing Slide-seqV2 data of kidney tissue by our collaborators Teia Noel,
@qbw_128
,
@AnnaGreka
, and
@JamieLMars
!
0
22
108
Did you bookmark our recent Slide-tags preprint but not get around to reading it? Very relatable
Fortunately for you, you can hear our very own
@jacksonweir4
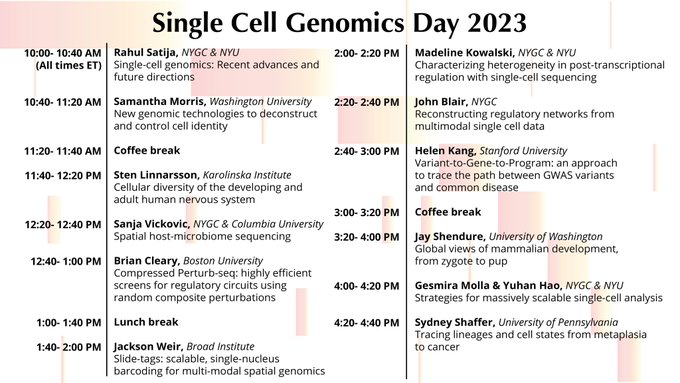
give a talk about it at Single Cell Genomics Day tomorrow at 1:40 ET!
3
7
56
Fantastic read on single-cell multi-omics featuring
@hattaca
providing insight into inCITE-seq
0
2
46
Our new method, C-SIDE, for cell type-specific DE in spatial tx:
Detects DE in diverse contexts such as space, disease, cellular interactions. Controls for cell type effects and mixing. Led by Dylan Cable and colab w/
@rafalab
@macosko
@Vignesh_Shan
0
6
37
In case you missed it, check out our new method for photoselective sequencing, led by
@s_mangiameli
!
Sharing Photoselective Sequencing (PSS), a new method for genomic and epigenomic measurements within spatial regions of interest.
Thanks to the whole team
@haiqi_chen
,
@H3K4me1
,
@juliedobkin_
,
@DLesman
,
@JD_Buenrostro
, and Fei Chen.
(1/9).
1
43
112
0
1
17
On the other hand, we're thrilled to have new faces Gio, Julie,
@dawnchenx
,
@VerenaVolf
, Mehdi, Yifan,
@Luyi_T
, Simon, Chen, &
@AndyRusss
!
Looking forward to more exciting science and lab fun in 2022! Happy New Year!
0
1
11
This wouldn't be possible without two stellar teams of scientists and our ongoing collaborations with
@macosko
's group!
1
1
10
It was great to be invited to talk about Slide-tags!
Slide-tags: Combining single-nucleus genomics with spatial measurements 🧬🔬
Hear from
@AndyRusss
,
@jacksonweir4
, Naeem Nadaf, and
@insitubiology
as they describe the development and application of this impressive technology
0
2
15
0
0
10
Send us your technology papers!
Call for papers for our new special issue 'New technology applications in health and disease' guest edited by Francisco De La Vega
@ribozyme
& Fei Chen
@insitubiology
!
Deadline 8/31/22
#SingleCell
#spatialbiology
#artificialintelligence
#MachineLearning
3
8
12
0
2
9
10. Not a paper, but possibly even more exciting: in December,
@soph_liu
was awarded an
#ImpetusGrant
with Karin Gustafsson from the Scadden lab to study immune cell aging
Thanks to
#ImpetusGrants
@MartinBJensen
@LNuzhna
for funding our project! Grateful for the support from my PI,
@insitubiology
, and excited to work alongside Karin Gustafsson in David Scadden's lab.
1
2
22
1
1
7
Fast forward to December, we published two Nature papers, both released on the same day!
Congrats to
@AndyRusss
,
@jacksonweir4
, and
@Naeemnadaff
for Slide-tags and to Jonah and Nina for the spatial mouse brain atlas!
Excited to share two new papers, published today in
@Nature
!
Check them out here:
(1) Slide-tags, a new method to unite single-cell and spatial profiling:
4
99
370
1
0
7
With success comes people moving on, and it was bittersweet to see
@Andrew_C_Payne
(
@E11BIO
),
@PaulReginato
(climate tech), Julia Morriss (Harvard CCB), Bob Stickels (Scale Bio), Evan Murray,
@JamieLMars
(Solid Bio), &
@haiqi_chen
(UTSW) off to new adventures
2
1
7
6. In October,
@Luyi_T
outlined a vision for the future of tissues genomics in a comprehensive technology review (w/
@macosko
)
Luyi is starting his own group at Guangzhou Lab, be on the lookout for openings!
Our review on Spatial Transcriptomics is out
@NatureBiotech
! With
@insitubiology
and
@macosko
we reviewed the technology advance in sequencing and imaging-based ST and envisioned the future of tissue genomic where dynamics and perturbation measured at spatial levels.
3
228
785
1
0
6
11. And right before the holidays, Tongtong Zhao and
@z_chiang
adapted Slide-seq beads to measure DNA and used this technique to study clonal heterogeneity in cancer (with
@JD_Buenrostro
)
Just released
@Nature
: slide-DNA-seq, our new method for spatial DNA sequencing in tissues 🧬
Excited to add a genetic dimension to the rapidly expanding spatial toolbox with the tech dev superteam of Tongtong Zhao,
@JD_Buenrostro
&
@insitubiology
(1/6)
12
221
659
1
1
6
8. In October,
@JamieLMars
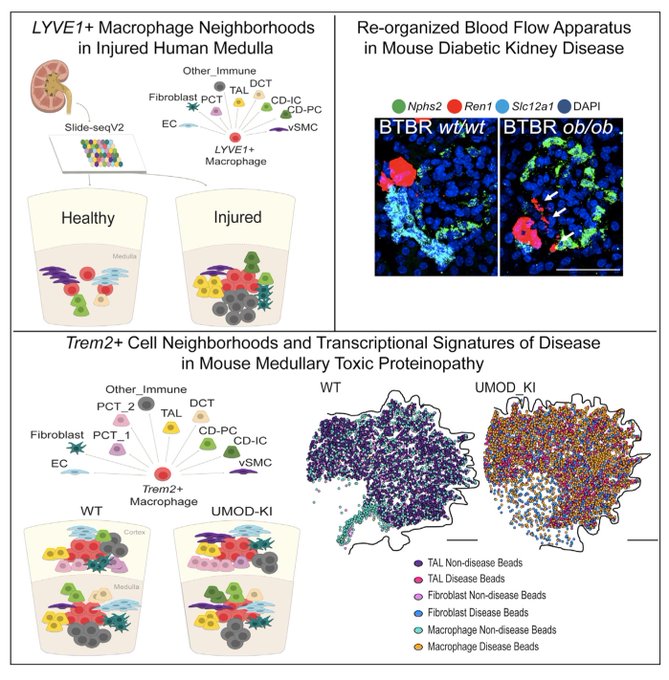
shared her work using Slide-seq to map cell neighborhoods in healthy and diseased kidneys
With Teia Noel,
@qbw_128
,
@SilvanaBazua
,
@haiqi_chen
, &
@AnnaGreka
Thrilled to share the Kidney Slide-seq pre-print . HUGE thanks to the amazing kidney slide-seq team Teia Noel,
@qbw_128
, and
@haiqi_chen
. Thanks to Evan Murray for the Slide-seq arrays,
@SilvanaBazua
, and
@AnnaGreka
, Fei Chen, and
@macosko
for mentorship!
1
5
34
1
1
6
7. In June,
@ehshabibi
and collaborator
@DanielaJDiBella
released a single-cell atlas of mouse corticogenesis (supervised by Aviv Regev &
@ArlottaLab
)
Very excited to share my paper just out in
@NATURE
.
KUDOS to
@DanielaJDiBella
from
@ArlottaLab
, Aviv, Fei, Bob Stickels, Gabriele Scalia, and Tommasso Biancalani for the heroic effort!
14
54
269
1
1
6
In July,
@Luyi_T
's work alongside
@AbhiSampKumar
and Adriano spatially mapping mouse organogenesis came out in Nature Genetics!
1
0
6
4. In February, Dylan Cable published RCTD, a computational approach for deconvolving cell type mixtures in spatial transcriptomics data (with
@rafalab
)
1
2
6
2022 brought so much new energy with our group moving into a new space and welcoming
@HuChenlei
,
@jacksonweir4
,
@sundakao
,
@SandeepKambham2
,
@benno_orr
,
@WollensakDavid
, &
@RuthRaichur
We also celebrated
@juliedobkin_
&
@Luyi_T
taking their next steps!
Started at the bottom (
@broadinstitute
10th floor), now we're here (Broad 11th floor) 🚀
Celebrating the opening of our new lab space with a DIY ribbon cutting ceremony
9
6
403
1
0
5
In April,
@s_mangiameli
’s new method for genomic and epigenomic measurements within spatial regions of interest, Photoselective Sequencing (PSS), was published in Nature Methods!
Photoselective Sequencing is out in
@naturemethods
! This was a collaborative effort between the
@insitubiology
and
@JD_Buenrostro
labs, and I couldn't have done it without my co-authors
@haiqi_chen
,
@H3K4me1
,
@juliedobkin_
, and
@DLesman
!
1
11
43
1
0
5
We saw the graduation of our first two Chen lab PhDs.
Dylan is completing a post-doc at Stanford and will be starting his own lab at UMichigan soon!
Sophia is now leading the Liu Lab at the Ragon Institute as an Early Independence Fellow!
The Ragon is proud to welcome our newest Early Independence Fellow, Sophia Liu (
@immunoliugy
), joining Sept 1. Her lab will focus on tools to study cell interactions for enhanced understanding of the immune system.
Welcome, Dr. Liu!
#RagonInstitute
#Immunology
#WomenInSTEM
2
10
146
1
0
5
6. In April, collaborator Brian Cleary published Composite In Situ Imaging, a compressed sensing method for imaging transcriptomics
With Aviv Regev &
@eric_lander
+ contributions from Evan M,
@sinhanubhav
,
@ehshabibi
, &
@JamieLMars
1
1
5
8. Lastly, in 2022 we started to really see the results of our work to make spatial tech more broadly accessible -- a true team effort led by Evan Murray and
@Irving_Barrera1
1
0
5
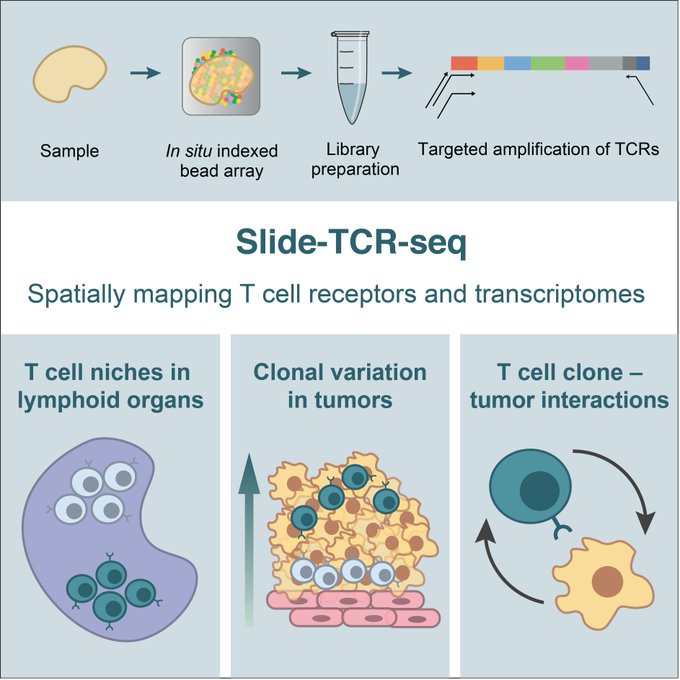
7. A week later,
@soph_liu
brought together spatial tech and immunology with Slide-TCR-seq, a method for spatially mapping T cell receptors and transcriptomes
With Bryan Iorgulescu, Shuqiang Li, and Catherine Wu
Presenting Slide-TCR-seq: our method for measuring the transcriptomes of cells & T cell receptor (TCR) sequences in situ!
This collaboration w/ Bryan Iorgulescu, Shuqiang Li, Catherine Wu, and
@insitubiology
led to some cool findings...
1/8
19
206
808
1
0
4
9. In November,
@haiqi_chen
released the first spatial transcriptomic atlas of mammalian spermatogenesis
Haiqi is starting his own lab
@UTSWNews
and is recruiting at all levels!
Excited to share our work published today
@CellReports
on dissecting spermatogenesis using spatial transcriptomics.
2
21
55
1
1
4
We are very excited to have you
@jacksonweir4
!
Very excited to share I have joined the Chen Lab
@insitubiology
for my PhD! I will develop new spatial omics tools to answer pressing questions in tissue dynamics and cancer evolution! 🔬🧬
7
4
87
0
1
4
1. We kicked off 2021 with in situ genome sequencing, a new technique for measuring DNA sequence and structure in intact samples
With
@Andrew_C_Payne
,
@z_chiang
,
@PaulReginato
,
@eboyden3
, &
@JD_Buenrostro
Introducing IN SITU GENOME SEQUENCING, a new technology to resolve DNA sequence🧬 and structure🔬
A team effort with
@Andrew_C_Payne
,
@PaulReginato
,
@eboyden3
,
@JD_Buenrostro
, and Fei Chen
This is how it works: (1/8)
13
577
1K
1
1
4
Do you use computational methods to solve important questions at the intersection of
#biology
,
#informatics
, and
#medicine
? HSCRB and
@HarvardDBMI
are jointly recruiting for a tenure-track faculty. Learn more and apply by 12/1/22.
0
1
4
@Irving_Barrera1
started medical school at Harvard, Simon started graduate school in New York,
@WollensakDavid
moved to Johns Hopkins to start his PhD, Diane joined the Duke MD-PhD program, and
@anlau19
began a research position at Stanford!
1
0
4
1. In January, we introduced RADARS, a new programmable way to target cells using RNA sensing
With
@idmjky
,
@jgkoob
,
@dawnchenx
,
@RohanKrajeski
, Yifan Zhang,
@jgooten
, and
@omarabudayyeh
Today we report in
@biorxiv
programmable RNA sensors that express any protein gated on specific RNAs in a cell. Much like gene sequencing necessitated gene editing tools, we answer “What is the Cas9 of single cells?” 🧵👇🏼w
@jgooten
&
@insitubiology
1/17
9
162
648
1
1
4
3.
@hattaca
dropped inCITE-seq for jointly measuring nuclear proteins and RNA in single cells, which enables quantifying the direct or indirect effects of TFs on gene expression in vivo (supervised by Aviv Regev)
1
1
3
We were also excited to welcome new scientists to the lab:
@doralidsy
and Yunhao started as post-docs,
@kevin921tw
and Nick joined as PhD students, Mehdi began as a senior computational associate,
@yupines
and DK joined as RAs, and Larissa and Matthew started as undergrads.
1
0
3
Or if you prefer,
@omarabudayyeh
's:
Today we report in
@biorxiv
programmable RNA sensors that express any protein gated on specific RNAs in a cell. Much like gene sequencing necessitated gene editing tools, we answer “What is the Cas9 of single cells?” 🧵👇🏼w
@jgooten
&
@insitubiology
1/17
9
162
648
0
0
3
2. ExSeq was published just weeks later, combining in situ transcriptomics and expansion microscopy
With Shahar Alon,
@D_R_Goodwin
,
@sinhanubhav
, Oz Wassie,
@geochurch
,
@AdamMarblestone
, &
@eboyden3
1
1
3
3. March saw the publication of
@JamieLMars
's work on the spatial organization of kidney disease
With Teia Noel,
@qbw_128
,
@haiqi_chen
, and
@AnnaGreka
🎉 Greka lab paper alert! Our team is excited to share a new story published in
@iScience_CP
led by
@JamieLMars
describing the use of Slide-seqV2 to identify disease-specific cell neighborhoods in human and mouse kidneys.
5
17
82
1
0
3
5. In March,
@SGRodriques
&
@linlinmc
’s work on RNA timestamps to encode the age of single molecules was released in print (with
@eboyden3
)
New paper out today in
@NatureBiotech
with Linlin Chen, Fei Chen,
@eboyden3
, and many other wonderful coauthors, demonstrating how we can encode the age of an RNA into its sequence to achieve "temporal transcriptomics"
11
160
548
1
1
3
4. Jumping to August,
@hattaca
shared snFFPE-seq, paving the way for single nucleus RNA-seq of archival clinical samples
With Alexandre Melnikov, Cristin McCabe, and Aviv Regev
1
0
2
5. In September,
@s_mangiameli
introduced photoselective sequencing (PSS), which uses fluorescence imaging to make precise spatial genomic measurements (w/
@JD_Buenrostro
)
Sharing Photoselective Sequencing (PSS), a new method for genomic and epigenomic measurements within spatial regions of interest.
Thanks to the whole team
@haiqi_chen
,
@H3K4me1
,
@juliedobkin_
,
@DLesman
,
@JD_Buenrostro
, and Fei Chen.
(1/9).
1
43
112
1
1
2
@dawnchenx
and
@SandeepKambham2
got exciting news about fellowships:
Dawn was awarded the AHA Predoctoral Fellowship and Sandeep was awarded an NSF GRFP!
1
0
2
2. In February, Dylan Cable followed up his popular RCTD method with C-SIDE, enabling differential expression for spatial transcriptomics (w/
@rafalab
)
Our new method, C-SIDE, for cell type-specific DE in spatial tx:
Detects DE in diverse contexts such as space, disease, cellular interactions. Controls for cell type effects and mixing. Led by Dylan Cable and colab w/
@rafalab
@macosko
@Vignesh_Shan
0
6
37
1
0
1
tweetorials on the bioRxiv from
@jgooten
@omarabudayyeh
linked here:
Genome editing owes a huge debt to genome sequencing
With efforts like
@humancellatlas
, we’re sequencing cells like mad. What is the "Cas9" of single cell?
With
@omarabudayyeh
@insitubiology
: RADARS, for programmable cell-specific sensing/expression 🧵👇
8
100
494
0
0
1
Check out
@jgooten
's tweetorial for more details:
Genome editing owes a huge debt to genome sequencing
With efforts like
@humancellatlas
, we’re sequencing cells like mad. What is the "Cas9" of single cell?
With
@omarabudayyeh
@insitubiology
: RADARS, for programmable cell-specific sensing/expression 🧵👇
8
100
494
1
0
1
The tweetorial by
@idmjky
describes our updates since our bioRxiv:
1
0
1
Here's hoping for another great year of science and lab fun in 2023!
Still in a reminiscing mood? Check out our 2021 year in review:
0
0
1