Rahul Satija
@satijalab
Followers
28K
Following
841
Media
172
Statuses
900
Core Member, New York Genome Center; Professor, Biology, NYU
Joined September 2009
Interested in 3' UTR changes in scRNA-seq? Check out our: * CPA-Perturb-seq paper * UCSC Perturb-seq tracks (see who regulates your favorite 3’ UTR!) * Seurat extension for polyA site analysis (PASTA) * Deep neural perturbation network (APARENT-Perturb) https://t.co/XXYEBHV1iG
cell.com
CRISPR perturbation screens of 42 cleavage and polyadenylation regulators followed by 3′ scRNA-seq (CPA-Perturb-seq) identifies modules of co-regulated polyA sites, their connection with distinct...
5
89
312
The peer-reviewed version of expansion in situ genome sequencing is now out in Science! The news is bittersweet – when we first revealed this last September, I never guessed it would be my final paper in academia, but a lot has changed. A few parting thoughts:
18
299
2K
Excited to share our first anti-aging intervention study, led by brilliant Zehao @Tommyz626, Alex @alexepstein_bio, and Chloe @chloe__schaefer from our lab at @RockefellerUniv! With a cost-effective spatiotemporal analysis pipeline, we identified the aging-associated cell
Why do many aging interventions come & go, yet Caloric Restriction (CR) remains a robust pillar, extending healthspan across species? Its benefits are even noted in ancient eastern texts like The Yellow Emperor’s Classic of Internal Medicine!
5
20
87
I'm thrilled to share our new #bioRxiv preprint from @junyue_cao lab at @RockefellerUniv! We present PerturbFate, a high-throughput single-cell CRISPR platform profiling nascent/pre-existing RNA, steady-state RNA & chromatin accessibility.
biorxiv.org
High-throughput genomic studies have uncovered associations between diverse genetic alterations and disease phenotypes; however, elucidating how perturbations in functionally disparate genes give...
8
42
306
Our Single Cell Genomics Day - highlighting our favorite new sc/spatial methods - starts today at 10AM ET! Talks are free and live-streamed (no registration required) on Youtube and https://t.co/zG98gkckMU
7
37
179
Our Single Cell Genomics Day - highlighting our favorite new sc/spatial methods - starts Friday at 10AM ET! The most common question we get: is this open to anyone? Yes! Talks are free and live-streamed (no registration required) on Youtube and https://t.co/zG98gkckMU
3
88
303
Excited to share our exploration @naturemethods into new horizons opening up in the era of low cost sequencing! 🔥 Deeper sequencing at WGS scale for PPM sensitivity of tumor informed MRD 🔥Duplex sequencing at WGS scale for plasma only exploration of ctDNA Check out thread 👇
What happens when sequencing costs go ⏬ ? Imagination and new opportunities go ⏫ @landau_lab Let's look at what you can do when you perform deep WGS on cfDNA for cancer detection. Our recent work with the @UltimaGenomics platform 🧵👇 Lets goooo!! https://t.co/ZMaJqUKNDK
3
24
116
Interested in single cell genomics but need help getting started? Check out the full agenda for our Single Cell Genomics Day on Friday 4/25. All talks will be live-streamed (no registration required) at https://t.co/JqaO6urVPF
4
205
587
We're incredibly excited to share our new method, Paired-Damage-seq, out @naturemethods today! This technique allows us to jointly analyze oxidative DNA damage and SSBs with RNA, all at the single-cell level (1/5).
nature.com
Nature Methods - This work presents an in situ labeling-by-repair strategy, Paired-Damage-seq, to achieve joint profiling of oxidative and single-stranded DNA damage alongside gene expression in...
4
20
68
Thanks to our wonderful keynote speakers @WJGreenleaf , @HongkuiZeng , and @jure - with many more to come soon!
0
0
3
Interested in single cell genomics but need help getting started? Check out my lab's Single Cell Genomics Day on April 25. Talks will feature recent exciting computational and experimental advances and will be live-streamed at https://t.co/zG98gkckMU. Please RT/spread the word!
10
241
564
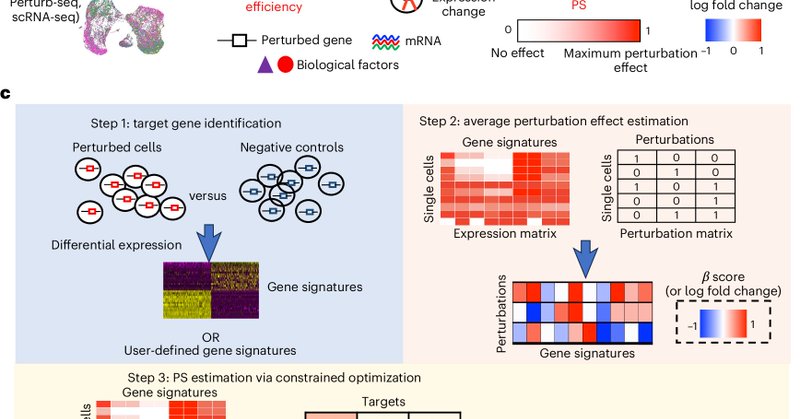
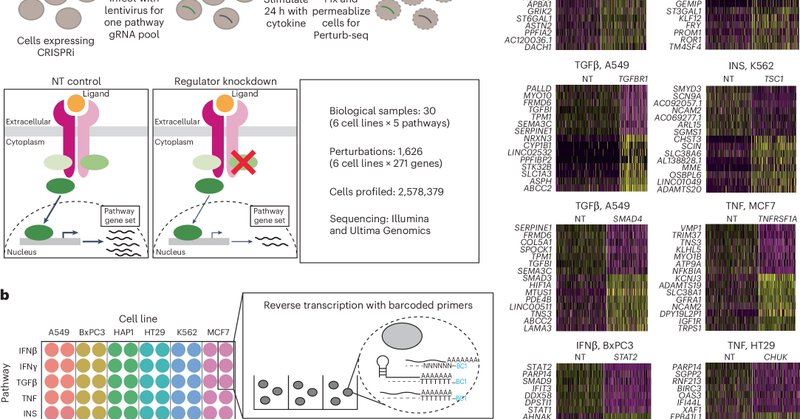
And congrats to @davidliwei lab for their co-published computational method to analyze heterogeneous Perturb-seq data!
nature.com
Nature Cell Biology - In two independent studies, Song et al. and Jiang, Dalgarno et al. present computational frameworks, perturbation-response score and Mixscale, respectively, to determine...
0
1
15
Papers led by incredible lab members John Blair @Longda_Jiang @carol_dalgarno We're using these methods in many of our projects now, and hope you will find them useful in your own work! Many thanks to our generous funders @_hubmap @cziscience and @genome_gov via @cegs_ica
1
2
9
Check out our two new papers with new single-cell tech/methods/data! 1. Phospho-seq: Multi-modal profiling of intracellualar proteins https://t.co/6z5rx0UeNU 2. Systematic Perturb-seq of signaling regulators (2.6M cells, 6 cell lines, 1500 perturbations) https://t.co/PPc31sW0dO
nature.com
Nature Cell Biology - In two independent studies, Song et al. and Jiang, Dalgarno et al. present computational frameworks, perturbation-response score and Mixscale, respectively, to determine...
3
86
348
Excited to share a major breakthrough from the lab! D&D-seq is a new technology that bridges a key gap in single-cell multi-omics/genomics : the ability to map DNA:Protein (e.g., TFs or chromatin remodellers). Led by Ivan Raimondi, Wei-Yu Chi and Sang-Ho Yoon
🧬 We’re excited to introduce D&D-seq, a single-cell technology that maps DNA:Protein interactions through molecular footprinting.. Check it out here: https://t.co/mfsDO3kKa0
#Genomics #Epigenetics
9
49
241
Finally out in @ScienceMagazine! Utilizing the cost-effective EasySci to profile 21 million single-cell transcriptomes across five life stages and diverse sexes/genotypes, we reveal aging as distinct, development-like transitions, with dramatic cell population changes in specific
What if 1 million scRNA-seq libraries can be prepared within $1k? Here we go 👉 21 million cells from 623 mouse tissues, spanning 5 life stages & 3 genotypes, all in a SINGLE study by ONE remarkable student Zehao @Tommyz626 from our lab @RockefellerUniv!
11
60
277
In collaboration with Reuben Saunders, @JswLab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! https://t.co/iJ8hi3ddz4
biorxiv.org
Organ function requires coordinated activities of thousands of genes in distinct, spatially organized cell types. Understanding the basis of emergent tissue function requires approaches to dissect...
2
116
341
Incredibly excited that our paper linking single molecule states of TF binding to gene expression using quantitative thermodynamic models is out in Nature today. An amazing collaboration with the Bintu Lab. Congrats to Ben, Michaela, and Julia!
nature.com
Nature - A study uses single-molecule footprinting to measure protein occupancy at regulatory elements on individual molecules in human cells and describes how different properties of transcription...
10
108
514
Cool work from @spyros_darmanis that performs combinatorial-based superloading of @10xGenomics for scRNA-seq and multiome assays. Interesting idea to add the 10x barcode first, then the second barcode later:
0
21
82
We are hiring for a wetlab research association position! Check it out to learn more about our lab's focus in single-cell genomics, multimodal/spatial analysis, and genome engineering. Apply/learn more at https://t.co/nrK32zxsvn or DM me a copy of your CV!
jobs.silkroad.com
Find a career with New York Genome Center
2
26
50
humbled to announce KdT IV, an oversubscribed $100M+ fund dedicated to supporting early-stage, science-driven companies. at KdT, we believe the most iconic companies of our time are those that leverage scientific breakthroughs to build enduring businesses with significant
29
11
272