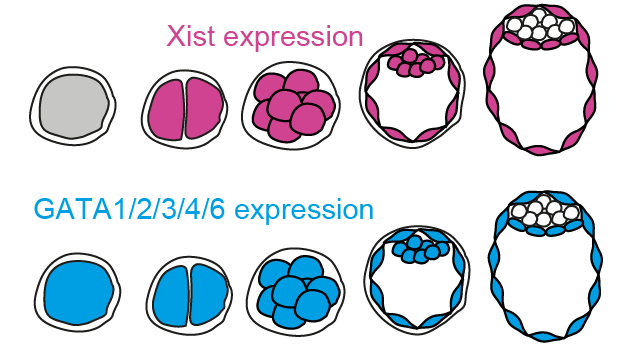Abhishek Sampath Kumar
@AbhiSampKumar
Followers
418
Following
3K
Media
6
Statuses
178
Developmental epigenetics | Postdoctoral fellow @ArlottaLab @Harvard | https://t.co/QB5WND7Yb6…
Cambridge, Massachusetts , US
Joined June 2019
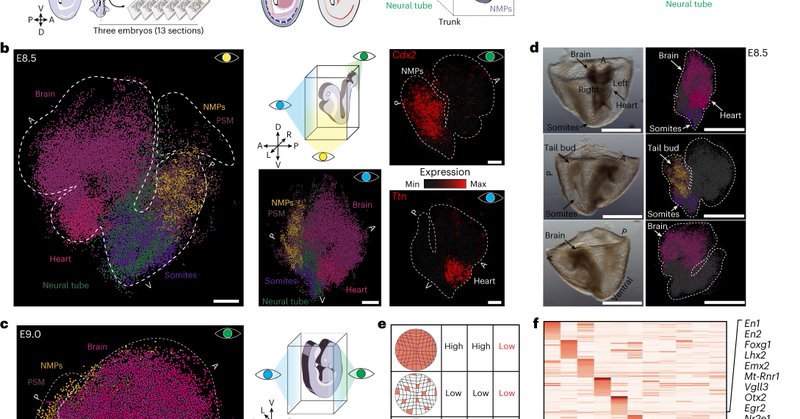
Excited to share the second piece of my PhD work! Our paper on generating 2D and 3D spatial transcriptomic maps of whole mouse embryos during early organogenesis with @adrianobolondi @Luyi_T is out in @NatureGenet 🐭🧬 @MPI_MolGen @broadinstitute.
nature.com
Nature Genetics - Slide-seq profiling of mouse embryos at the onset of organogenesis (embryonic days 8.5–9.5) coupled with a new three-dimensional reconstruction and visualization tool (sc3D)...
17
87
359
Check out this amazing work from @PGuckelberger @jengreitz , answering an important question on how 3D contacts tune enhancer effects on gene expression. Congratulations to the entire team.
I am so excited to share this part of my PhD where we tackle the question: What impact do 3D contacts have on how enhancers regulate their target genes? . 1/
1
0
8
First one out from @Luyi_T lab in @naturemethods congratulations to all authors. Happy to have contributed.
Our revised paper is online at @naturemethods !! the revised version contains 11 methods with more analysis. We keep updating our data portal ( so it contains recent technologies that not in the paper, such as Visium HD.
0
0
11
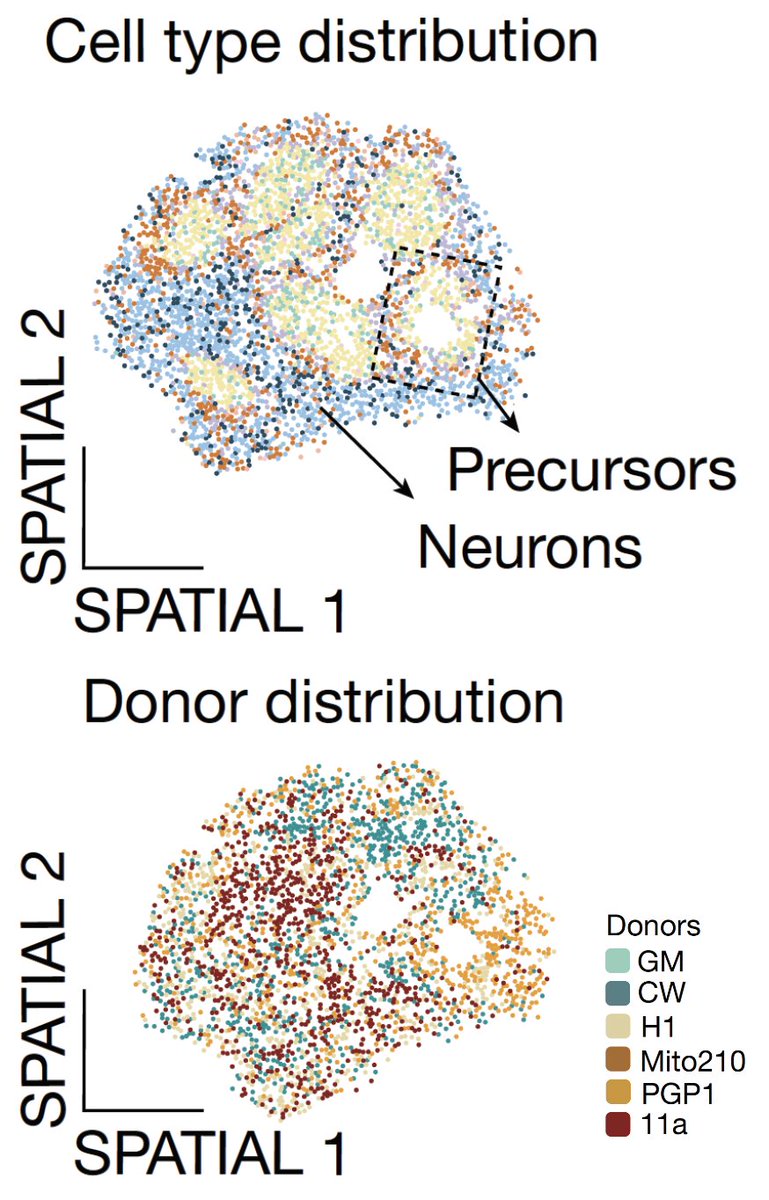
In the multi-donor brain chimeroids, we also show that the different donor lines are spatially distributed, profiled with Curioseeker @curiobio . Excited to have contributed to this tremendous work led by @BolanosAnton @IreneFaravelli @ArlottaLab. Congratulations to all authors.
📢Thrilled! My 1st piece of work of my postdoc at @Arlottalab is out @Nature! This has been a huge co-lead effort with @IreneFaravelli👩🔬👩🔬🤩 .Link: Tweetorial 👇🧵
0
3
22
A fantastic and systematic study from @Maxistoe @bulutkarslioglu ! Kudos to @Maxistoe for his hardwork, dedication and success. Happy to have contributed. Congratulations to all the authors @MPI_MolGen.
How do embryos and ESCs safeguard pluripotency throughout a state of dormancy? A great collaborative effort of the @bulutkarslioglu -lab & the.@MPI_MolGen involving @helenekretzmer @AbhiSampKumar @_taktas_ @VvdWeijden & many more.
0
0
9
I am excited to share the final version of this work published today in @NatureCellBio. A great collaboration with Liat,@EddaGSchulz and the entire team @MPI_MolGen.
We’re excited to share our new preprint, headed by Liat Ravid Lustig and @AbhiSampKumar, where we identify the GATA family as tissue-specific activators of Xist, which are required for initiation of X inactivation in early embryos. See tweetorial below
2
3
46
Register now to attend the webinar on the 1st of November. We’ll discuss our efforts in reconstructing 3D spatial maps and using it to understand tissue patterning in the embryo.
Join our webinar and learn about experimental and computational framework for the spatiotemporal investigation of whole embryonic structures and mutant phenotypes. Register today:
0
2
12
Join us on the 1st of November to learn about our efforts in reconstructing 3D spatial transcriptomic maps of whole mouse embryos.
Join our webinar and learn about experimental and computational framework for the spatiotemporal investigation of whole embryonic structures and mutant phenotypes. Register today:
0
0
17
Check out this really exciting work from @ArlottaLab @BolanosAnton @Irene_Faravelli and team. The multi-donor chimeroid with various genetic backgrounds is a great platform for scalable testing of genetic susceptibility to neurotoxic triggers.
📢📢Today our 🧠 cortical Chimeroid model is out in @biorxivpreprint 👀. Very excited to share the first piece of work from my postdoc in @Arlottalab. This has been a huge co-lead effort with @Irene_Faravelli. 👩🔬👩🔬🤩. See the “tweetorial” below 👇👇.
0
0
8
RT @HCRimaging: This week’s #FluorescenceFriday user is Abhishek Sampath Kumar! @AbhiSampKumar applied #HCRimaging to confirm the expressio….
nature.com
Nature Genetics - Slide-seq profiling of mouse embryos at the onset of organogenesis (embryonic days 8.5–9.5) coupled with a new three-dimensional reconstruction and visualization tool (sc3D)...
0
3
0
Check out how sc3D can be used to reconstruct, visualize and analyze 3D spatial gene expression patterns from other datasets (such as Stereo-seq) @GuignardLab @TheBgiGroup.
I was wondering if sc3D was able to register and display the crazy data from Here is the result with some Hox genes🤓 (showing downsampled data). sc3D➡️➡️@AbhiSampKumar, @adrianobolondi, @Luyi_T et al.:
0
1
26
RT @HSCRB: New research in @NatureGenet presents an experimental and computational framework for the spatiotemporal investigation of whole….
nature.com
Nature Genetics - Slide-seq profiling of mouse embryos at the onset of organogenesis (embryonic days 8.5–9.5) coupled with a new three-dimensional reconstruction and visualization tool (sc3D)...
0
4
0
RT @adrianobolondi: 🚨Paper alert🚨 With @AbhiSampKumar, @Luyi_T & @GuignardLab we built a 3D spatial transcriptomic atlas of mouse embryos t….
nature.com
Nature Genetics - Slide-seq profiling of mouse embryos at the onset of organogenesis (embryonic days 8.5–9.5) coupled with a new three-dimensional reconstruction and visualization tool (sc3D)...
0
10
0
RT @GuignardLab: 📢📢Always wanted to look at all expressed genes in whole E8.5/9.0 mouse embryos, at near single cell resolution while keepi….
0
13
0
Big shout out to @AAragonesH , Bob Stickels, Evan, @helenekretzmer , Maria, Lars, Leah, Gabriel, @ElkabetzYechiel ,@macosko ,@insitubiology ,@GuignardLab , and Alex for their remarkable contributions to this work.Your collaboration has been instrumental in this endeavor. (11/11).
1
0
5
We find that 'ectopic tubes' display a conflicting molecular identity (neural and mesodermal) and loss of complete DV patterning. This highlights an uncoupling between morphogenetic outcomes and transcriptional state. #NotInTheGenes (9/11).
1
0
2