Evan Macosko
@macosko
Followers
3K
Following
204
Media
5
Statuses
78
Genomics technologist, Psychiatrist, Associate Professor, Massachusetts General Hospital @broadinstitute. Founder, Curio Bioscience
Joined May 2012
We developed an imaging-free spatial genomics technology where DNA barcodes diffuse to connect locally. Using UMAP, we reconstructed the physical locations of these barcodes, transforming spatial transcriptomics into purely molecular biology. https://t.co/BIPlyibPXa
biorxiv.org
Tissue organization arises from the coordinated molecular programs of cells. Spatial genomics maps cells and their molecular programs within the spatial context of tissues. However, current methods...
6
64
256
How do genetic perturbations change cells? How are these effects shaped by cell type and dosage? How do we best extract insight from modern massive perturbation atlases? Im pleased to share a new preprint where we develop a suite of statistical approaches to these Qs (link below)
9
77
296
Hear us talk about Slide-tags!
Slide-tags: Combining single-nucleus genomics with spatial measurements 🧬🔬 Hear from @AndyRusss, @jacksonweir4, Naeem Nadaf, and @insitubiology as they describe the development and application of this impressive technology https://t.co/K6iYlKwuFy
0
4
23
I am thrilled to announce that I will be joining the Department of Genetics at Trinity College Dublin (@tcdgenmicro) in summer 2024! My lab will build and apply new tools to understand the formation and function of microglia states in brain health and disease.
18
21
248
Findings "strongly imply that there is no specific link between maternal mental illness or maternal infection during pregnancy" & neurodevelopmental disabilities (including autism) in offspring https://t.co/ja8N4C0Cp7 free
cambridge.org
The causal association between maternal depression, anxiety, and infection in pregnancy and neurodevelopmental disorders among 410 461 children: a population study using quasi-negative control...
4
19
38
A huge congratulations to Jonah Langlieb, Nina Sachdev, @Naeemnadaff, @KarolBalderrama, Mukund Raj, and Chuck Vanderburg. And grateful for the continued collaboration with @insitubiology We hope (and expect!) our atlas helps accelerate many areas of neuroscience research.
0
0
11
We now know the locations of cell types in a full mammalian brain. Our work reveals the brainstem’s absolutely stunning cellular complexity. How and why does it arise? Visit https://t.co/vlGT8w8EO7 to plot genes, localize types, and get markers. https://t.co/VOtqEZvkSO
2
87
314
Slide-tags unifies the single cell + spatial genomics worlds. It solves three key problems with existing spatial methods: 1) Cell segmentation (the biggest problem) 2) robustness and scalability (well-powered case-control studies now possible) 3) Multiomics (ATAC, RNA, etc)
A new method called Slide-tags lets scientists capture both genetic and location information of individual cells using standard single-cell workflows in the lab. The technology builds upon Slide-seq, both developed by the labs of @insitubiology & @macosko. https://t.co/2LsxjxZa9L
3
42
276
The sharing of data is the only way we will make collective progress on hard problems like AD. Our entire dataset and analysis are available at https://t.co/9OkNhULxKh Vahid Gazestani’s integration of 27 datasets can be easily imported to answer your own scientific questions.
3
3
25
Utilizing rare human brain biopsies we address: 1) Which cell types likely make amyloid? 2) How do microglia respond to amyloid and how does this compare to states in mouse models? 3) What neuronal states precede cell death in AD? Paper out today! https://t.co/KhrWb3wjPk
cell.com
Generating single-nucleus atlas from cortical biopsies of living individuals at early stage of Alzheimer’s disease, cell states of neurons, microglia, and oligodendrocytes associated with AD pathol...
7
50
222
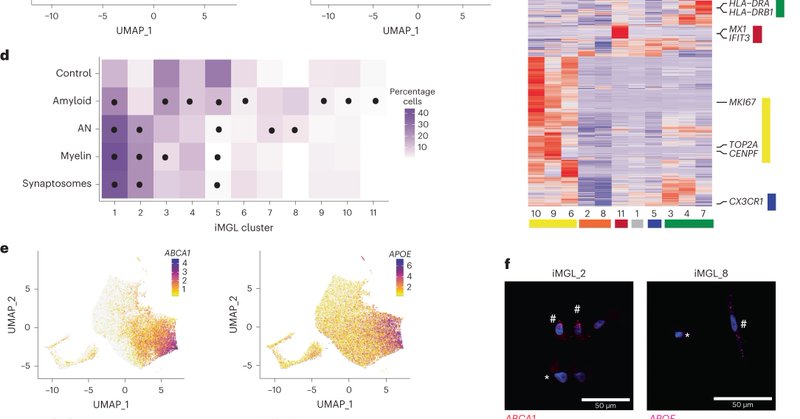
Delighted to announce our work developing an approach to understand the function of human microglial states. @MartineTherrie4 and I will guide you through our work, a collaboration with @sasajereb from @macosko and @stevens1lab !
nature.com
Nature Immunology - Stevens and colleagues show that human stem-cell-differentiated microglia can be used to model the extensive transcriptional diversity of human brain microglia.
11
46
158
Congrats @tkam80, Vahid Gazestani and dear collaborators Ville Leinonen and @LabMalm. Amazingly, Vahid integrated our data with most postmortem datasets: https://t.co/eS0Z2JPrV7 We hope his work provides a common language for discussing cell types and states in human cortex.
0
3
14
How does brain tissue change when amyloid first appears in the brain? In rare surgical biopsies, we find a hyperactive neuronal state preceding loss, and evidence of oligodendrocytes, as well as neurons, producing amyloid. https://t.co/R9a9lUnzNz
2
31
146
Single-cell or spatial? Our new technology - Slide-tags - allows both in the same experiment, enabling true single-cell multi-modal spatial genomics ➡️ https://t.co/i1m5T4bEme
12
168
582
All single cell assays can now be spatial. Slide tags is simple, reliable single cell with hi-res spatial positioning. A truly outstanding scientific team: @insitubiology, @andyrusss, @jacksonweir4, Naeem Nadaf. https://t.co/X5r5TSktz6
biorxiv.org
Recent technological innovations have enabled the high-throughput quantification of gene expression and epigenetic regulation within individual cells, transforming our understanding of how complex...
1
59
229
Introducing Slide-tags: our new technology for truly single-cell, multi-modal spatial genomics! Had a blast working alongside @AndyRusss, @Naeemnadaff, @macosko, @insitubiology, and the rest of our team on this project! https://t.co/WXav5awW9u 1/9
6
24
131
Example uses of https://t.co/vlGT8w9cDF: 1) label transfer your sc data to ours. Get cell type assignments and spatial positions. 2) Find genes or cell types enriched in a region. 3) Retrieve the minimal set of genes needed to define a cell type uniquely (ie for FISH).
0
0
12
An amazing team led by Jonah Langlieb and Nina Sachdev. Kudos to Karol Balderrama, who did Slide-seq on an entire brain in just three (!) days, and Naeem Nadaf, who collected 6 million (!) snRNA-seq profiles. Always grateful for our ongoing collaboration w/ @insitubiology.
0
0
5
Two of my broad conclusions from these data: 1) The midbrain + medulla deserve far more attention in neuroscience than they currently receive; 2) Functionally annotating all of these cell types will require new tools, perhaps building on the RADARS/RADAR/CellREADR technologies.
2
0
11
Excited to release our cell type atlas of the adult mouse brain! Here’s our portal: https://t.co/9OkNhUM5zP And preprint: https://t.co/4FhnGfBBsU We find minimal marker sets for all cell types, explore activity-regulated genes, and perform analyses of heritability enrichment.
biorxiv.org
The function of the mammalian brain relies upon the specification and spatial positioning of diversely specialized cell types. Yet, the molecular identities of the cell types, and their positions...
2
51
213