Luke Koblan
@LukeKoblan
Followers
661
Following
730
Media
16
Statuses
161
K99/R00 fellow at @WhiteheadInst in @JswLab | Helen Hay Whitney Fellow | Chemical Biology PhD @Harvard in @liugroup
Joined January 2017
RT @WhiteheadInst: PEtracer combines lineage tracing and spatial data, providing a detailed view of how cancer growth is influenced by both….
news.mit.edu
Whitehead Institute researchers developed a tool to recreate cells’ family trees. Comparing cells’ lineages and spatial data or cells’ location within a tumor provided insights into environmental and...
0
1
0
I'll be out at Stanford this coming Thursday hosted by my friend and collaborator @weallen1 where I'll be presenting a seminar on our recent PEtracer work ( Feel free to swing by-- info attached!
0
5
115
Excited to announce that this work is now live as a first release article @ScienceMagazine So grateful for this fantatic team and to see this work in wild!
science.org
Charting the spatiotemporal dynamics of cell fate determination in development and disease is a long-standing objective in biology. Here we present the design, development, and extensive validation...
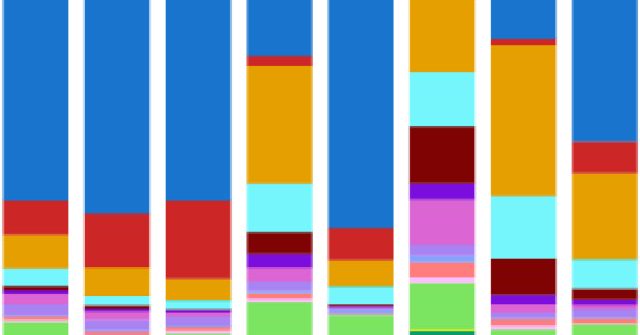
1/14.On behalf of the amazing team in @JswLab, we’re excited to share PEtracer ( a prime editing-based evolving lineage recorder compatible with both scRNA-seq and high-resolution imaging readouts in intact tissue. By applying PEtracer in a syngeneic mouse.
6
19
89
There are of course tons of other cool findings in the paper and we hope you give it a read! This has been an incredible collaboration with @KatieEYost, @sclerei, @WilliamNColgan, as well as our co-authors (@MattG_Jones, @dianyang1117, @alexa_schnell, @sundakao, @_canergen) and.
2
0
5
6/14.Building off of elegant work from the @ElowitzLab @AskaryLab for in situ barcode readouts (see PMID: 31740838) , we perform extensive tissue digestion and clearing before amplifying lineage tracing cassettes using T7 polymerase. We then decode integration barcodes and edits
1
0
1
1/14.On behalf of the amazing team in @JswLab, we’re excited to share PEtracer ( a prime editing-based evolving lineage recorder compatible with both scRNA-seq and high-resolution imaging readouts in intact tissue. By applying PEtracer in a syngeneic mouse.
biorxiv.org
Charting the spatiotemporal dynamics of cell fate determination in development and disease is a long-standing objective in biology. Here we present the design, development, and extensive validation...
3
57
191