Jasper Janssens
@JasperJanssens7
Followers
433
Following
571
Media
16
Statuses
132
Postdoc @TreutleinLab @ETH_en EMBO fellow | PhD @ Aerts lab | KU Leuven @CBD_VIB | Fruit fly enthusiast -- Gene regulation -- Brain development #EvoDevo He/him
Joined December 2017
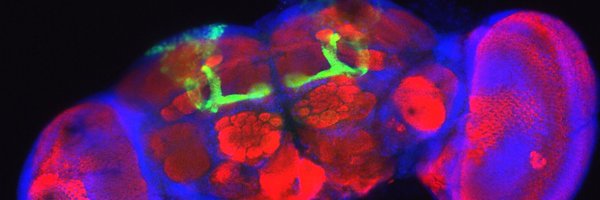
Our paper describing gene regulation in the fly brain is out now in @Nature! So proud of this one and of the amazing team work with @SaraAibar @ibrahimihsan and @joynismail in the @steinaerts lab! Check it out here https://t.co/gkFcgDtLKX with below our main findings:
14
150
621
Our ChromBPNet preprint out! Huge congrats to @panushri25! This was quite a slog for both of us but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Link in the next tweet. Bluetorial coming soon .. 1/
1
13
45
Big day for neuroscience & the end of the beginning for whole brain connectomics. Shout out to @sdorkenw and Philipp Schlegel, first authors of the connectome papers, @MurthyLab @SebastianSeung for establishing #flywire to make this possible and @dddavi for birthing this dataset.
Big news! The fly connectome is featured on the cover of a special edition of Nature. This is all possible thanks to the collaboration of 292 members of The FlyWire Consortium! https://t.co/Mrz7Yrx1eU Check out the thread for an overview of the 9 #flywire papers published today
6
53
237
Happy to share CREsted, our latest package on enhancer modeling! CREsted allows for training sequence-based deep learning models from scATAC-seq data, enhancer decoding and enhancer design through a variety of methods. Check out the GitHub here: https://t.co/hesNelFprs. [1/9]
1
41
129
Last week I got awarded the @ASLeuven award for Biomedical Sciences @KU_Leuven for my PhD research 🪰🧠. Very proud and humbled to receive this award and eternally gratefull to all the amazing scientists who I was able to work with, especially my mentor @steinaerts! Thanks!!🙏🙏
🏆 The ASL Award for Biomedical Sciences (@KU_Leuven) was recently awarded to @JasperJanssens7—a @CBD_VIB alumnus of the @steinaerts lab— for his PhD research on decoding gene regulation in the fly brain 🪰🧠 We catch up with Jasper to find out more ➡️ https://t.co/k633aYRAIm
4
2
56
The fly has a full #connectome, basically a directed + weighted graph. But what are its connection ‘signs’, excitatory/inhibitory? We move towards this answer, predicting transmitters from EM! w/@BobQubit, vid @amyneurons, 1/n Published @CellCellPress: https://t.co/yuWZEVWZkx
5
106
376
Very excited to announce our new preprint together with Nikolai Hecker! Dive into our findings on the conserved regulatory codes shaping cell identity across species. 🧬 [1/n] https://t.co/l0eCJXTTZ6
biorxiv.org
Combinations of transcription factors govern the identity of cell types, which is reflected by enhancer codes in cis-regulatory genomic regions. Cell type-specific enhancer codes at nucleotide-level...
1
22
64
Do you love big data and the brain? We have #ScienceJobs for you! In Drosophila there are >11,000 cell types that define the circuit structure of the connectome – but we don't know their molecular identity. Integrating connectomes and transcriptomes would be revolutionary. RT 1/3
1
42
57
How much of our genome is 'noise' and how much is actually functional? In the de Boer lab's latest paper (along with @DarkMatterDNA) we aim to uncover the truth behind this long standing debate! 1/ https://t.co/68Pyt7JAPK
https://t.co/Iis47rVz9Y
4
49
174
Thrilled to share my PhD work in the @TreutleinLab & @GrayCampLab! In this 🚨preprint🚨, we present an integrated perspective on how morphogens impact early human neural organoid development: https://t.co/yueNr6ULWb Tweetorial landing in 3, 2, 1...
5
30
148
Happy to announce the first paper of my postdoc in the @TreutleinLab! A fun collaboration with @HsiuChuan_Lin where we investigated how morphogen signaling pathways can be used to increase the diversity of TF-induced neurons. Check the paper here: https://t.co/ESPXwSOIF0!
biorxiv.org
Human neurons programmed through transcription factor (TF) overexpression model neuronal differentiation and neurological diseases. However, programming specific neuron types remains challenging....
Programming diverse neuron subtypes from stem cells! Coupling patterning with proneural TF induction, we screened for 480 morphogen signaling modulations in ~700K cells. Great work with @JasperJanssens7 @TreutleinLab @GrayCampLab
https://t.co/8PGJrJdz3E
7
16
123
Very happy to see my final PhD paper on designing synthetic enhancers for selected tissues in vivo using #AI out in @Nature 🥳 Big thanks to @AlexanderStark8 and whole @stark_lab , as well as collaborators in @Eileen_Furlong lab! Check out our thread below for more details 👇
We are very excited to present a major breakthrough achievement – the de novo design of synthetic enhancers for selected tissues in fruit fly embryos in vivo using deep- and transfer learning, @deAlmeida_BPet al published today in @Nature
https://t.co/TIlPrBKYaK. Thread 👇(1/N)
7
9
118
Exciting news! Our latest research on "Cell type directed design of synthetic enhancers" has been published in @Nature. We've harnessed deep learning to design enhancers with cell-type specificity. Find our paper here: https://t.co/wC44fDMp70. Highlights below: [1/n]
22
255
956
We are recruiting! If you are interested in neuronal evolution or neuronal circuits or you just enjoy generating new tools in different bugs, please consider applying. If not, please retweet :-) More information by email or DM. You can apply here: https://t.co/DSqTwSwHxr
0
96
107
Extremely happy to be awarded an #ERCSyG grant @ETH_en @ETH_BSSE together with Elly Tanaka @Tanaxolotl @IMPvienna & Kevin Briggman @MPINB_outreach! In #AxoBrain, we will map cell types, architecture & circuits of the axolotl brain and explore its amazing regenerative capacity!
📣 Results of the 2023 ERC Synergy Grant competition are out! 37 research groups win a total of €395 million in #ERCSyG funding 👉 This will enable them to pool diverse expertise and resources to push the frontiers of knowledge. Full story: https://t.co/qxp34JIn4u 🇪🇺#EUfunded
7
11
129
Human brain atlas papers by @kimsiletti @MossiAlejandro @ka_wai_rine
@ErnestArenasSWE @EmelieBraun1 @MiriDanan Rebecca Hodge Trygve Bakken Ed Lein @LarsBorm Elin Vinsland Erik Sundström and many others. With a stunning double cover illustration by @IKapustova
In 21 papers across Science, @ScienceAdvances, and @ScienceTM, researchers with the #BICCN consortium present an atlas of the human and nonhuman primate brain in unprecedented detail. The results uncover the genetic organization of the human brain. https://t.co/K3ePBMRzli
2
67
248
36 datasets, 26 protocols, 1.7 million harmonized cells! First edition Human Neural Organoid Cell Atlas. Important work from @zhisong_he @josch_f Leander Dony @TreutleinLab @fabian_theis. Strong community effort @Sergiu_P_Pasca @GiorgiaQuadrato. #organoids @ETH_BSSE @IHB_Research
6
160
669
Spatial transcriptomics in adult flies! I feel rejuvenated by finding localised mRNAs in gigantic flight muscles, 20 years after studying bicoid mRNA in oocytes. Fantastic collaboration with @steinaerts @JasperJanssens7 @PierreMangeol & co-thank you all! https://t.co/aYRmcHGywn
6
40
206
Massive thanks to @PierreMangeol, @KatinaSpanier and @joynismail for creating beautiful slices. To @gapartel, @NikolaiHecker, and @PierreMangeol for analysis and to @SchnorrerL and @steinaerts for mentoring. A fantastic and fun collaboration and very happy with the results!!! 🥳
1
0
3
and mapped unknown scRNA-seq clusters to spatial locations. Check out the paper for all details and analysis!
1
1
3