Gray Camp
@GrayCampLab
Followers
4K
Following
2K
Media
140
Statuses
686
Curiosity. Human Model Systems. EvoDevo. CompBio. Translation. Institute of Human Biology. Basel. [email protected]
Joined May 2013
What makes our cells human? The evolution of human development is undeniably fascinating. It is an exciting time to explore the details! We, @brainevodevo, @craiglowe, @KlkUmut, @NatureRevGenet review and suggest new inroads: https://t.co/hncKPOCBxn
#sapiens #evodevo #organoids
4
44
246
PDAC is a terrible malady and needs new inroads. We integrate a cell atlas, develop 3D tumoroid models, and map phenotypic spaces at scale. Very cool data inside! @IHB
https://t.co/nxNkT69oKN,
https://t.co/I5txaxRhUv
2
19
69
I was at the @FCBasel1893 vs. Lugano game tonight and managed a great video of the follow-up from the superb penalty save...looks like a goal to me?! I didn't see a better perspective on highlights (@SRF?), so sharing it here! Hope it gets to the fans! 😀⚽️
6
1
3
We are searching for computational PhDs and Postdocs to join our group in collaboration with @TreutleinLab . We have exciting single-cell technology projects that incorporate bioengineering, computation, biology, translation. Send CV to deco_ihb@roche.com
7
97
404
Human-mouse cross-species comparison identifies common and unique aspects of intestinal mesenchyme development https://t.co/6EC6M7MVkP A companion piece to our recent work from my colleague Kate Walton, led by Kelli Johnson and Xiangning Dong!
Excited to share our latest preprint led by Kelli Johnson- Mapping mesenchymal diversity in the developing human intestine and organoids! We defined 5 mesenchymal populations occupying discrete anatomical locations within the human intestine. https://t.co/rzSfsdGhjO
0
6
16
We made the cover! 🎉 Our vascular organoid paper is featured on the August issue of @CellStemCell Stem Cell. Huge shoutout to Liyan Gong for dreaming up the cover concept. Always exciting to see our science come to life visually. https://t.co/Tnd6bLvYIC
11
19
119
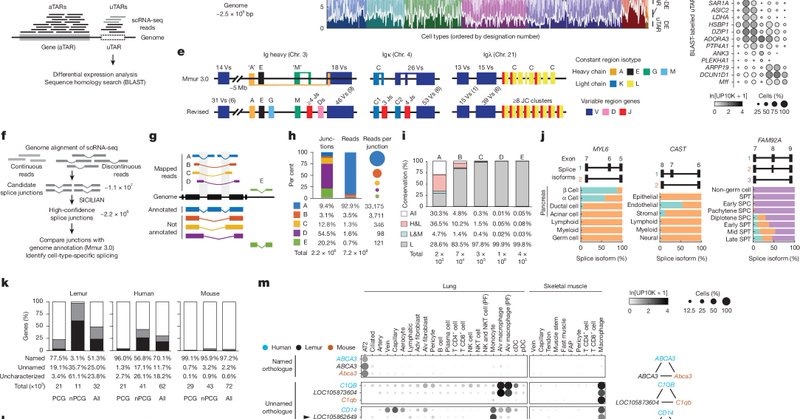
From Krasnow, Quake, and the Tabula Microcebus consortium @Nature A molecular cell atlas of mouse lemur: https://t.co/q9y4c8OJ16 Atlas informs primate genes, physiology and disease: https://t.co/mkja7SU90F News and Views:
nature.com
Nature - Together with an accompanying paper presenting a transcriptomic atlas of the mouse lemur, interrogation of the atlas provides a rich body of data to support the use of the organism as a...
0
6
25
Director at Max Planck - a unique position! The Open Call for Expressions of Interest in Max Planck Directorships is open now and can be submitted by the 31st of October 2025. ➡️ https://t.co/kz0macEMTX - Please share the Open Call among potential candidates.
4
50
100
Excited to share our latest preprint led by Kelli Johnson- Mapping mesenchymal diversity in the developing human intestine and organoids! We defined 5 mesenchymal populations occupying discrete anatomical locations within the human intestine. https://t.co/rzSfsdGhjO
2
25
121
We are happy to announce an opening for a Tenure Track Assistant Professor Faculty Position in Neuroscience at EPFL. Join our groups working on cellular & circuit neuroscience & neurocomputation - https://t.co/sdbArRbpyu. Deadline Oct 1 2025, Apply now - https://t.co/ghLJVC5Tcn
0
28
72
@JimWellsLab @TheSpenceLab Altogether, we identify the developing human intestinal epithelium as a rapidly evolving system, and show that great ape #organoids provide insight into human biology. 🙏Thanks to all authors/reviewers! Especially co-first authors: @CY_Qianhui_YU @KlkUmut @stefanosecchia 11/11
0
2
5
@JimWellsLab @TheSpenceLab We identify and functionally evaluate 2 human accelerated regions (HARs) at the IGFBP2 locus. Provocatively, these data indicate that the IGF pathway contributed to evolutionary change in the developing human intestine, potentially augmenting small intestine length. 10/11
1
0
2
@JimWellsLab @TheSpenceLab Organoid KO/Reporter assays confirm a bona fide Lactase expression enhancer within an MCM6 intron. Deep learning predicts that a selected SNP associated with lactase persistence removes a transcriptional repressor binding site, a potential mechanism for hypolactasia. 9/11
1
0
0
@JimWellsLab @TheSpenceLab We used a composite annotation to identify intestinal gene regulatory regions that have strong signatures of function and evolutionary change in humans. Willow plots aesthetically showcase the diverse gene ontologies affected by regulatory changes. 8/10
1
0
1
@JimWellsLab @TheSpenceLab We identified conservation and divergence between human and chimpanzee intestinal epithelial gene regulation associated with metabolic and barrier functions. 7/10
1
0
1
Truly incredible chimpanzee intestinal tissues can be generated from chimpanzee pluripotent stem cells, providing a very exciting inroad into comparative species analysis! Builds on extraordinary work from @JimWellsLab @TheSpenceLab #organoids #evodevo 6/10
1
0
1
Pluripotent stem cells can generate intestinal organoid tissues that have remarkable similarity to primary tissue counterparts. We performed a multiplexed reporter assay to assess putative enhancers for activity in intestinal organoids. 5/10
1
0
0
We explored the evolution of cell types and gene regulation by comparing tissue and #organoids from humans, chimps, and mice. Many genes and reg. regions active in enterocytes and associated with barrier function and metabolism have signatures of recent change. 4/10
1
0
0
At birth, the human GI tract becomes exposed to the outside world. Prior to birth, gene regulatory networks must orchestrate intestinal development to prepare for dynamic diet and microbiota exposures, which change over lifespan. 3/10
1
0
0