Franco Izzo
@FrancoIzzo85
Followers
836
Following
3K
Media
14
Statuses
904
Assistant Professor at Icahn School of Medicine at Mount Sinai | Department of Oncological Sciences | Tisch Cancer Center || Opinions are my own
New York, USA
Joined November 2017
GoT-ChA! I am thrilled to announce that our paper "Mapping Genotypes to Chromatin Accessibility Profiles in Single Cells" is now out in @Nature !! 👩🔬👨🔬🧬If you want to link somatic mutations to epigenetic changes directly in patient samples: https://t.co/M9Y3wEvpuW 👇🧵
23
98
437
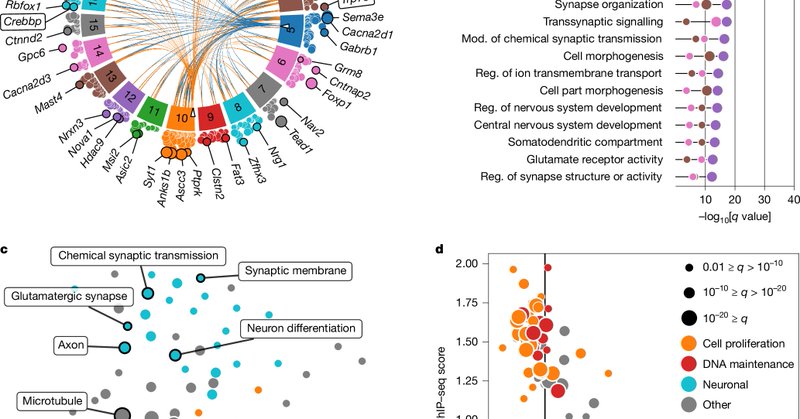
Nature research paper: Functional synapses between neurons and small cell lung cancer https://t.co/8OCZyKfqQD
nature.com
Nature - Small cell lung cancer cells form functional synapses with glutamatergic neurons, receiving synaptic transmissions and deriving a proliferative advantage from these interactions.
1
52
154
Our team is excited and honored to see our new method, HT SpaceM, for high-throughput single-cell metabolomics published in Cell! https://t.co/OEgNiEHgib 👇
5
51
208
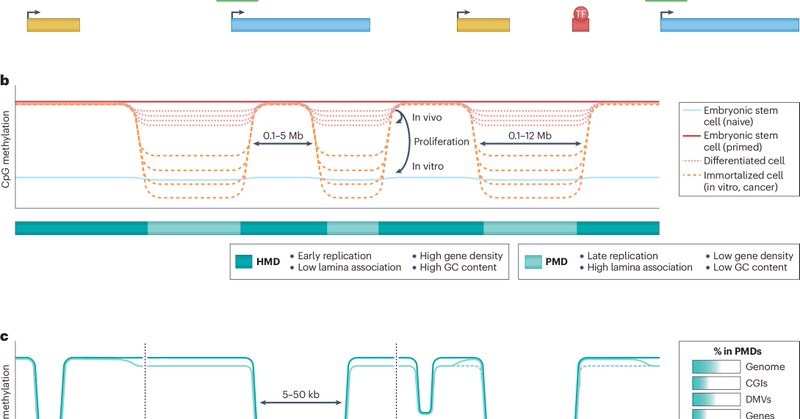
DNA methylation in mammalian development and disease https://t.co/XShWAoBisC
#Review by Zachary D. Smith @YaleMed, Sara Hetzel (@hetzel_s) & Alexander Meissner @MPI_MolGen Free to read here: https://t.co/MJWXDPXcyQ
@maxplanckpress
nature.com
Nature Reviews Genetics - In this Review, Smith et al. describe DNA methylation landscapes that emerge over mammalian development and within key disease states, as well as how different...
1
12
42
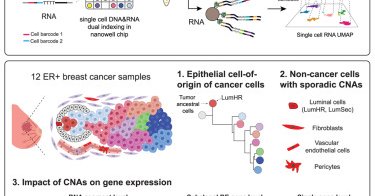
Amazing new paper from @nicholas_navin’s lab! Coalescing single-cell genomes and transcriptomes to decode breast cancer progression: Cell
cell.com
wellDR-seq allows single-cell whole-genome and transcriptome co-profiling at scale, enabling dissections of complex relationships between copy-number alterations and gene expression that inform our...
0
5
21
Excited to share our discovery of potent TRIM21 molecular glues with anticancer activity, online today @CD_AACR: "Defining the antitumor mechanism of action of a clinical-stage compound as a selective degrader of the nuclear pore complex". 1/18
4
31
122
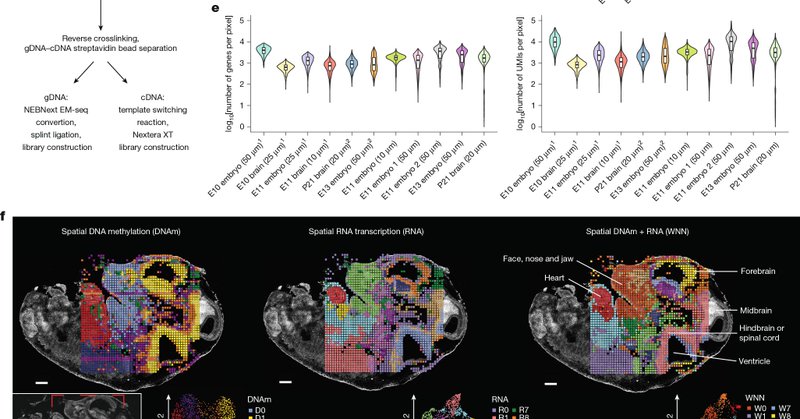
Spatial-DMT is now published in @Nature ! For the first time, we can simultaneously map DNA methylation and RNA transcription in tissue context. Great collab with @zhouwanding lab. Big congrats to Chin @Chin72155594 , Hongxiang @HongxiangFu and team!
nature.com
Nature - DNA-methylation and gene-expression profiling of tissue sections at near single-cell resolution can be used to create detailed spatial maps showing how methylation and transcription...
20
80
309
Nature methods: Co-profiling of in situ RNA-protein interactions and transcriptome in single cells and tissues https://t.co/P6GFcDcCfP
1
22
179
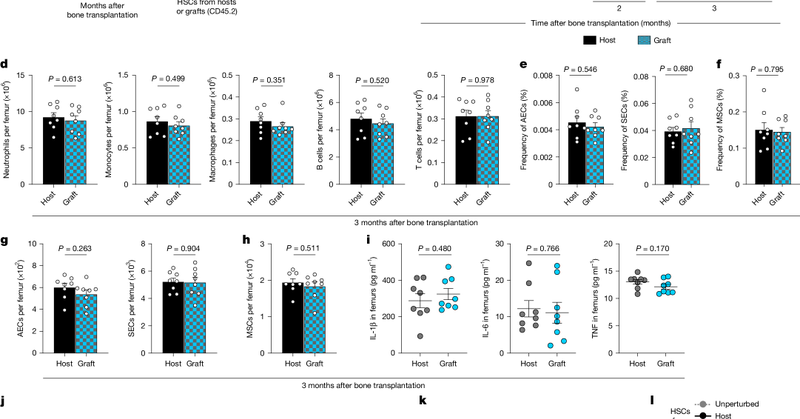
Excited to share our latest work @Nature! We show that the total number of hematopoietic stem cells (HSCs) in the body is restricted, challenging the classical model that HSC numbers are determined locally by the niche and introducing a new paradigm. (1/9) https://t.co/kvFqNQ8Nxb
nature.com
Nature - Haematopoietic stem cell numbers are restricted at both systemic and local levels.
7
64
251
A new bioRxiv preprint from the @landau_lab at New York Genome Center showcases the unmatched combination of sensitivity, efficiency, and cost-effectiveness made possible by #ppmSeq. Link to preprint: https://t.co/Ktk14kMfIm Link to PR: https://t.co/WBBdgweOwx
0
6
29
🧨Exciting new pre-print! Introducing ppmSeq - cost effective, scalable dual-strand 🧬 sequencing. Scalability & ultra-low error in one! A game changer for somatic genome discovery science and cfDNA clinical apps. Check out thread for deets! @AlexandrePCheng @UltimaGenomics
Highly accurate sequencing OR high throughput? We refuse to choose. Introducing paired plus-minus sequencing (ppmSeq), a method built to deliver both. Our latest paper with @UltimaGenomics 👀👀👀 See ⬇️ for a primer on the next frontier of genomics https://t.co/kl2KOaKziW
1
25
104
Highly accurate sequencing OR high throughput? We refuse to choose. Introducing paired plus-minus sequencing (ppmSeq), a method built to deliver both. Our latest paper with @UltimaGenomics 👀👀👀 See ⬇️ for a primer on the next frontier of genomics https://t.co/kl2KOaKziW
1
36
109
Researchers have developed a #DeepLearning system called BioEmu that rapidly generates diverse protein conformations, enabling fast, accurate insights into protein flexibility and function. Learn more this week in Science: https://t.co/Pe15hm9F52
17
214
719
Science doesn’t need to go according to plan; it just needs to lead to a discovery. If doesn’t have to be done alone or together with a buddy; there just needs to be a discovery. It doesn’t need to happen fast or slow; just as long as there’s a discovery, then everybody is happy.
15
266
1K
Recently published: Multimodal and Data-Driven Assessment of Myeloid Neoplasms Refines Classification across Disease States https://t.co/zhHhCQrS7I
0
4
19
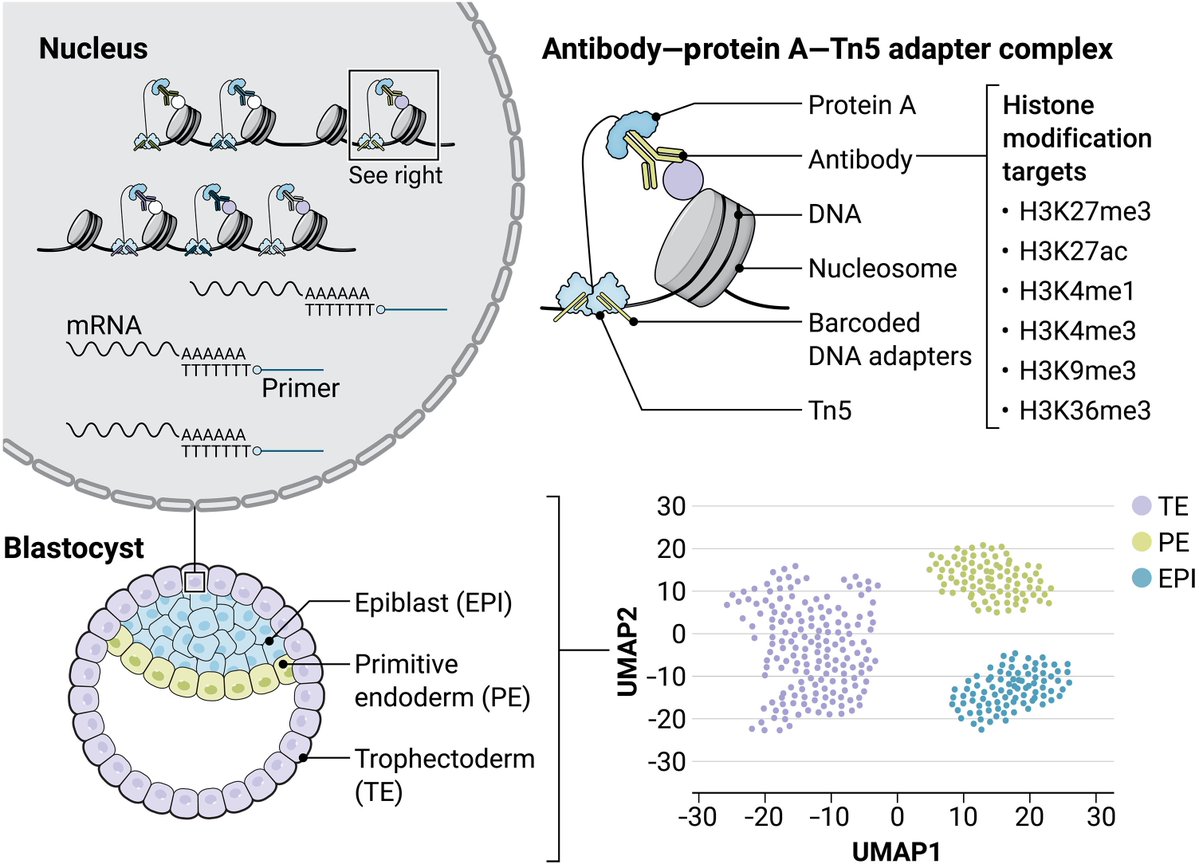
Single-cell multitargets and mRNA sequencing enable the simultaneous profiling of transcriptome and up to six histone modifications in individual cells @RuggGunnLab @ScienceAdvances
https://t.co/wpoUXLClye
https://t.co/xkOa8IQHzJ preview by @haiqi_chen
0
18
91
In GWAS studies, moving from a significant genomic locus to the driving genetic variants is challenging due to linkage disequilibrium. This review summarizes statistical fine-mapping methods that aim to detect potentially causal variants among sets of candidate variants.
2
35
166
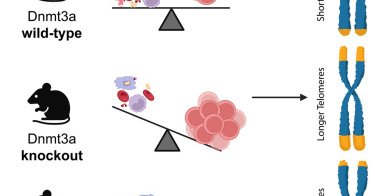
Online Now! Non-canonical functions of DNMT3A in hematopoietic stem cells regulate telomerase activity and genome integrity https://t.co/Twl44S84Ps
#stemcells
cell.com
Challen and colleagues show that the clonal expansion of Dnmt3a-null HSCs in serial bone marrow transplantation assays is not dependent on the loss of DNA methylation functions of Dnmt3a. They...
1
30
125
Enriching polygenic risk scores with single-cell ATAC-seq about chromatin accessibility in disease-relevant cell types shows potential to 👉improve predictive performance 👉better understand cells driving genetic predisposition to disease 👉uncover specific molecular drivers
1
15
74
For innate immune cells, the lack of diverse receptor rearrangements has prevented the study of clonal evolution of the innate immune system in human tissues and tumors. With @satpathology and @caleblareau, I’m delighted to share my PhD work that tackles this challenge:
Clonal lineage tracing of innate immune cells in human cancer https://t.co/uw2udDLVPp
#biorxiv_genomic
3
23
106
Time to mark our calendars moving for our 3rd "Probing Human Disease using Single-Cell Technologies" @Fusion_Conf taking place March 1-4, 2026 in beautiful Playa Mujeres, Mexico!! Our first 15 speakers are already on board and Early Bird and Talk submissions on Sept 3rd! Dont
Amy is looking forward to reuniting with familar faces, and welcoming new ones to the upcoming Single-Cell Technologies conference next year! A reminder that this meeting has sold out BOTH times, so do not delay getting registered and joining @iannisaifantis1 @satijalab and
0
7
14