Theodore Alexandrov
@thalexandrov
Followers
3K
Following
5K
Media
362
Statuses
2K
Head @alexandrovteam & Metabolomics Core @EMBL, BioStudio Faculty @BioCPH, @metaspace2020 lead, ERC Investigator, past co-founder https://t.co/gFwDTjxHAU, he/him/his
Germany / USA
Joined October 2009
Heidelberg is beautiful as always in any season 🌧
11
15
407
RT @BioinfoAdv: @thalexandrov 🧮 Access the tools and resources here: Web app: R package (S2IsoMEr): https://t.co/….
github.com
Spatial and Single cell Isomeric Metabolite enrichment in R - alexandrovteam/S2IsoMEr
0
1
0
RT @BioinfoAdv: @thalexandrov This paper introduces two complementary tools that address the challenge of ambiguous metabolite identificati….
0
2
0
RT @BioinfoAdv: 🧫 Now published in Bioinformatics Advances: “Enrichment analysis for spatial and single-cell metabolomics accounting for mo….
0
4
0
RT @slavov_n: The JPR issue on Single-Cell Omics is out. As the editorial highlights, understanding biological systems requires direct mea….
0
5
0
RT @metaspace2020: ⏳ Just one day to go! ⏳. Join us in our first webinar for insights on the latest features, best practices, and a live Q&….
eventbrite.com
Learn the top tips and tricks for identifying metabolites in Imaging MS using METASPACE - the ultimate guide for metabolite annotation!
0
2
0
RT @metaspace2020: 🔍 Did you know that you can select Regions of Interest (ROIs) in METASPACE?. We’ll be sharing more tips and best practic….
0
2
0
RT @metaspace2020: Ready to take your metabolite annotation to the next level?. Join our first FREE webinar on Best Practices for Metabolit….
eventbrite.com
Learn the top tips and tricks for identifying metabolites in Imaging MS using METASPACE - the ultimate guide for metabolite annotation!
0
3
0
RT @BrukerMassSpec: Join Bruker's upcoming webinar on Single-Cell Metabolomics: Revealing Cellular Diversity Using Isotope Tracing with Sha….
0
1
0
RT @FemalesInMS: 📢 Let us invite you to the ONLINE Career Transition Workshop with the amazing support of @scisteps 🙏. Link here for more….
0
7
0
RT @thalexandrov: Please welcome HT SpaceM, our new method for high-throughput small-molecule single-cell metabolomics: 👉Delafiori, Shahraz….
biorxiv.org
Single-cell metabolomics promises to resolve metabolic cellular heterogeneity, yet current methods struggle with detecting small molecules, throughput, and reproducibility. Addressing these gaps, we...
0
22
0
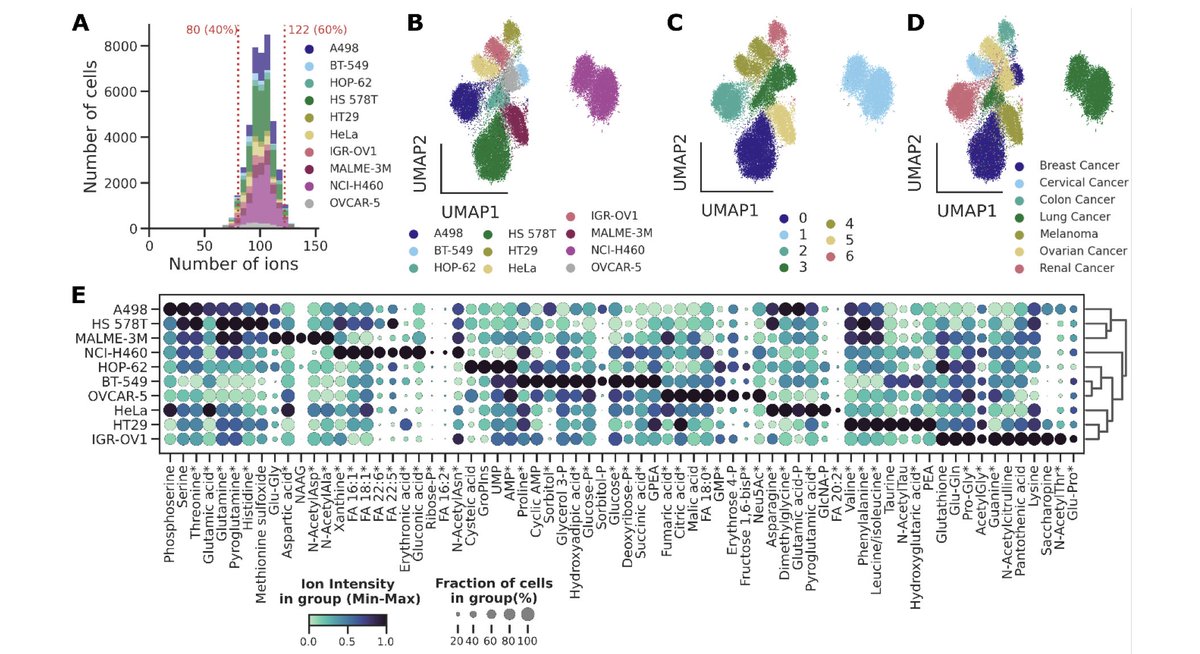
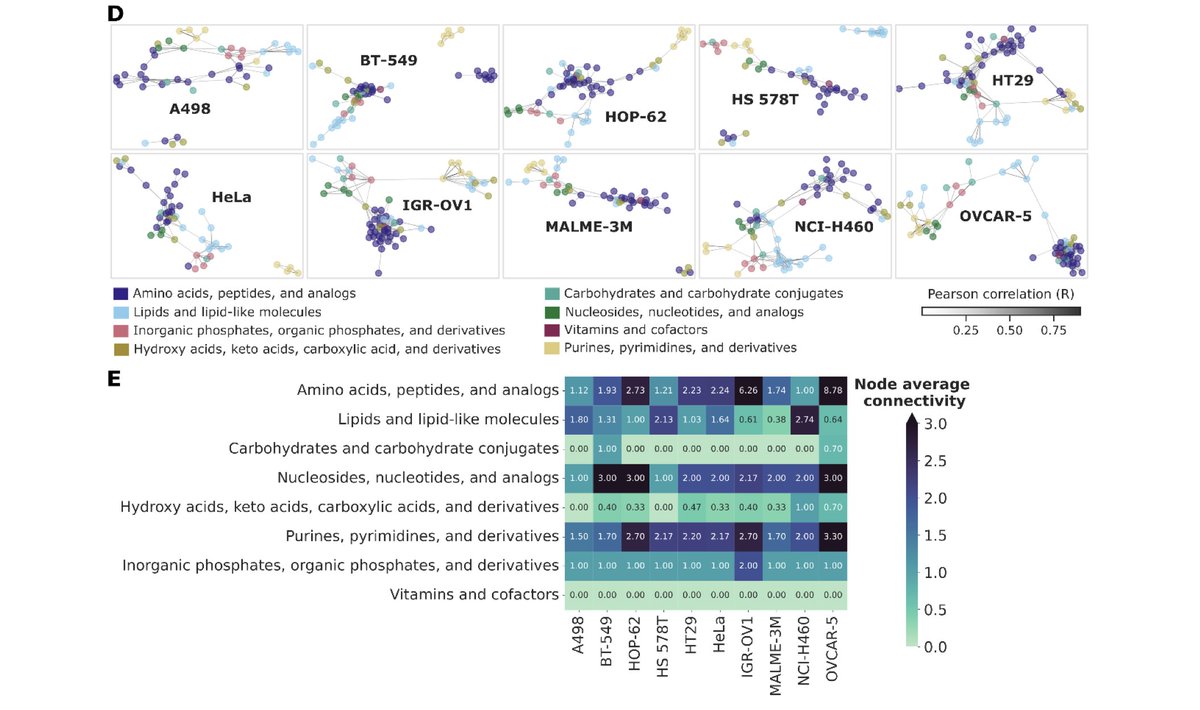
Please welcome HT SpaceM, our new method for high-throughput small-molecule single-cell metabolomics: 👉Delafiori, Shahraz et al, BioRxiv 🧵👇.
biorxiv.org
Single-cell metabolomics promises to resolve metabolic cellular heterogeneity, yet current methods struggle with detecting small molecules, throughput, and reproducibility. Addressing these gaps, we...
4
22
60