Vincent Liu
@vincentzliu
Followers
266
Following
285
Media
8
Statuses
68
Biotech enthusiast. Previously PhD in cancer immunology @stanfordmed and CS @columbia.
New York, NY
Joined May 2021
For innate immune cells, the lack of diverse receptor rearrangements has prevented the study of clonal evolution of the innate immune system in human tissues and tumors. With @satpathology and @caleblareau, I’m delighted to share my PhD work that tackles this challenge:.
Clonal lineage tracing of innate immune cells in human cancer #biorxiv_genomic.
3
23
104
@Satpathology @CalebLareau We think these findings suggest the tumor myeloid compartment may be peripherally programmed—raising the possibility of targeting peripheral monocytes to enrich DC-biased clones and boost anti-tumor immunity. Feedback is very welcomed!.
1
0
1
@Satpathology @CalebLareau Comparing TF activity in DC- vs mac-biased mono, IRF and NFkB factors stand out, highlighting the pro-inflammatory nature of this peripheral mono→tumor DC3 axis. These factors may prime mono for DC differentiation fate.
1
0
1
@Satpathology @CalebLareau Do mo-macs and DC3 share a common origin? We find clonally distinct mono subsets associate with each—DC- vs mac-biased mono exist in circulation w/ minimal overlap. Suggesting lineage fate is pre-set before tissue entry!
1
0
1
@Satpathology @CalebLareau Zooming into the myeloid compartment, we find tumor-enriched DC3s share epigenetic features of monocytes, and just like macs, they are highly related to both circulating and tissue-infiltrating monocytes, highlighting their peripheral origin.
1
0
0
@Satpathology @CalebLareau Myeloid cells across distant tissue sites are highly clonally related—pointing to continuous replenishment by circulating bone marrow–derived precursors, with minimal tissue-site bias.
1
1
2
@Satpathology @CalebLareau Lineage tracing revealed 3 major cell groups: 1) innate immune cells from recent hematopoiesis, 2) adaptive immune cells with clonal expansion, and 3) stromal cells of non-hematopoietic origin. Mono, DC, and mac were most closely related!
1
0
3
@Satpathology @CalebLareau From ~120k cells across 10 patients, we recovered ~10k clones—mapping their cell type and tissue distributions revealed 1) how clonal expansion varied across cell types and tissues, and 2) lineage relationships between cell types within and across sites.
1
0
2
@Satpathology @CalebLareau By leveraging somatic mtDNA mutations as barcodes, we used mtscATAC-seq to map lineage + epigenetic state of innate immune cells across tumors, normal tissue, and blood in pxts with NSCLC and ovarian cancer. Cell type–agnostic.
1
0
2
@Satpathology @CalebLareau While innate immune cells lack BCR/TCR rearrangements, their mitochondrial genomes (mtDNA) accumulate somatic mutations that are propagated during cell division, enabling clonal tracing. We used these mutations to track clonal evolution across human tissues and tumors.
1
0
3
RT @ScienceKyle: 🚨 New Preprint 🚨 .A continuous landscape of signaling encodes a corresponding landscape of CAR T cell phenotype. How can w….
0
57
0
RT @CalebLareau: Out today in @Nature, we describe HHV-6 reactivation in T cells, including therapeutic CAR T cells. I’m so grateful to hav….
0
118
0
RT @CalebLareau: Some big news: I’m absolutely delighted to announce that I will be starting my group this fall in the Computational and Sy….
clareaulab.com
The Lareau Laboratory for Computational and Systems Immunology is in the csBio Program at Memorial Sloan Kettering Cancer Center, New York, NY.
0
74
0
RT @dana_peer: Finally out, there are some papers that brings one real joy! Great collaboration with @CassBurdziak @LoweLabMSKCC and @Dir….
science.org
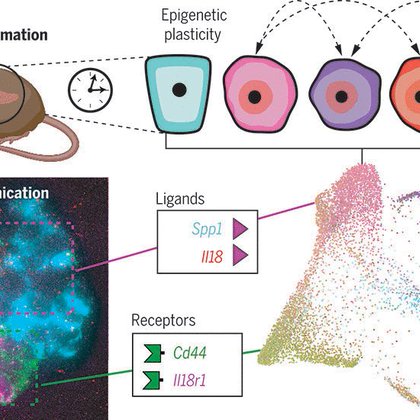
Single-cell analysis reveals that enhanced epigenetic plasticity drives pro-neoplastic cross-talk in early pancreatic cancer.
0
70
0
RT @CalebLareau: Out on @biorxivpreprint, I'm excited to share our latest deep dive into the world of mitochondrial genetics using single-c….
biorxiv.org
Somatic variation contributes to biological heterogeneity by modulating cellular proclivity to differentiate, expand, adapt, or die. While large-scale sequencing efforts have revealed the foundatio...
0
43
0
RT @joyp_ai: Excited to share that our work on regional T cell clonal responses to immune checkpoint blockade is out now! Very grateful to….
0
31
0
Proud to have contributed to this work!.
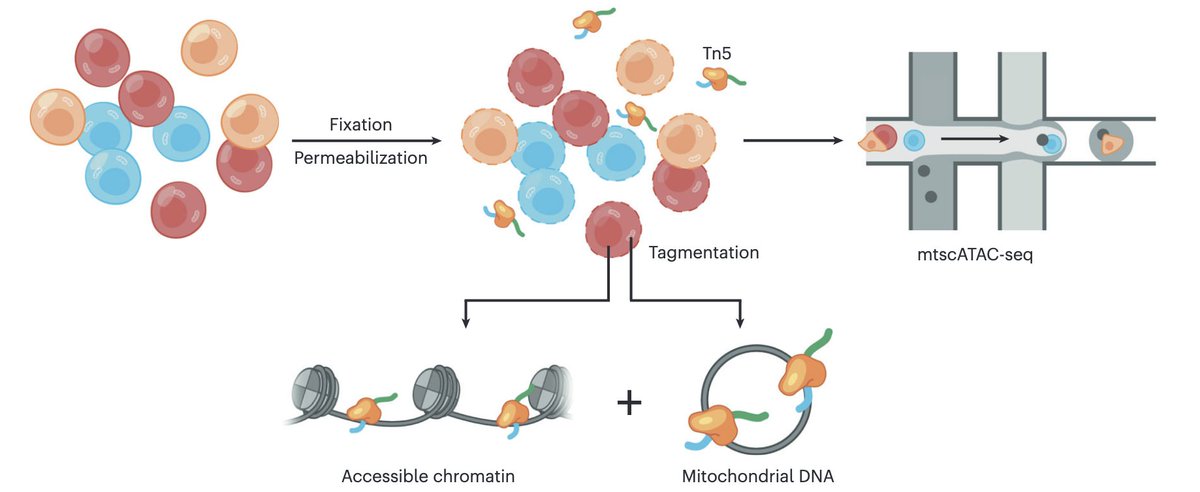
Out today in @NatureProtocols, our detailed writeup on the mitochondrial scATAC-seq assay, spanning everything from the molecular biology to computational analyses. Check out the paper here: and some highlights in this thread: (1/11)
0
0
12
RT @CalebLareau: Very inspired by this paper-- I don't think I've ever seen anything like this. I've never been able to lineage trace (wi….
0
18
0
RT @mollieblack418: Incredibly honored to present some of my postbac research next to other amazing scientists @sacnas in San Juan! have en….
0
7
0
Fascinating work dissecting the clonal dynamics of human NK cells with single-cell multiomics!.
We are excited to share our recent paper with the title “Clonal expansion and epigenetic inheritance shape long-lasting NK cell memory”, led by @TimoRckert, in which we study the origin and maintenance of human adaptive NK cells!
0
2
9