Matthew Loberg
@LobergMatthew
Followers
306
Following
610
Media
0
Statuses
267
Aspiring physician-scientist training @VanderbiltMSTP💊 | Interested in personalized cancer therapy🦀 | Part-time amateur blogger📝 | Opinions mine
Nashville, TN
Joined November 2018
@ltkagohara It was also great to see @TrowbridgeLab present all of the awesome work that her lab been doing uncovering mechanisms of clonal expansion and progression to leukemia in DNMT3a/NPM1c CHIP!!
0
0
1
I had so much fun presenting at AACR this week! Thanks to all of our collaborators for helping with this project! Including @ltkagohara for helping with the spatial data analysis.
Congratulations to @LobergMatthew for the awesome poster presentation at AACR. Thanks also to our fantastic collaborative team.
1
0
9
Congratulations to @LobergMatthew for the awesome poster presentation at AACR. Thanks also to our fantastic collaborative team.
0
3
16
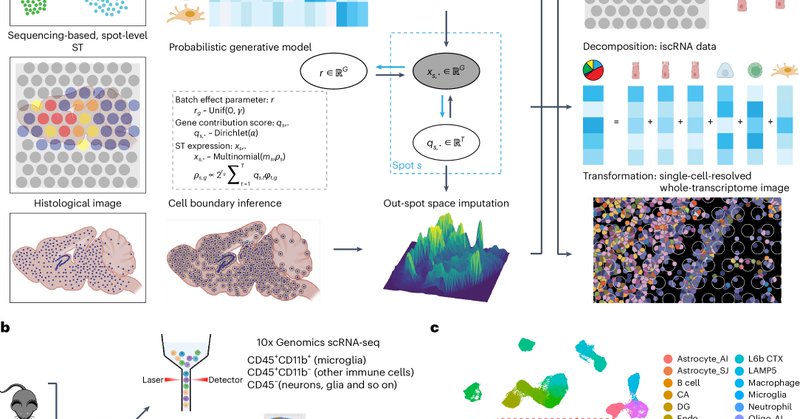
Spotiphy enables single-cell spatial whole transcriptomics across an entire section | Nature Methods
nature.com
Nature Methods - Spotiphy (spot imager with pseudo-single-cell-resolution histology) is a computational toolkit that transforms sequencing-based spatially resolved transcriptomics data into...
1
19
106
We are excited to share our new work out today in @NatureCellBio uncovering a new Achilles heel of pancreatic cancer. Myc is a key driver of PDAC and predicts poor survival. Thus, we asked the question of how Myc protein levels are regulated in PDAC. 👉 https://t.co/EMqDE5hX9M 🎉
4
9
41
We are excited to share our recent pre-print exploring cancer-associated fibroblast (CAF) phenotypes as thyroid cancers progress from treatable well-differentiated tumors to highly lethal anaplastic thyroid cancer! @DrVivianWeiss @hartheat21 @mtigue13
https://t.co/pXN21wICpK
biorxiv.org
Thyroid cancer progression from curable well-differentiated thyroid carcinoma to highly lethal anaplastic thyroid carcinoma is distinguished by tumor cell de-differentiation and recruitment of a...
1
2
11
Excited to share this preprint on TNC, Wnt, and thyroid cancer! Thanks @LobergMatthew for helping me with this!
Tenascin-C potentiates Wnt signaling in thyroid cancer https://t.co/YNNgWnVPsF
#biorxiv_cancer
0
2
9
Thrilled to share our back-to-back two Nature @Nature papers on the unexpected role of type 2 immunity against cancer. We found that interleukin 4 (Fc–IL-4), a type 2 cytokine, reinvigorates exhausted T cells to potentiate anticancer immunotherapy w/ @YugangGUO2 @RongFan8(1/11)
12
81
511
STAMP Single-Cell Transcriptomics Analysis & Multimodal Profiling through Sequencing-free, Imaging-based #SpatialOmics system CosMx Xenium Single-cell suspension cytospinned to slides 100 ~ >1M cells ⏫cell capture ⏬doublets Able to identify ultra-rare cell population
0
26
111
New preprint from @ltkagohara @FertigLab @DrLizJaffee @wonjinho1 @AshleyKiemen @AshaniTW & colleagues @hopkinskimmel @ConvergenceInst Spatial multi-omics reveal intratumoral humoral immunity niches associated with tertiary lymphoid structures in #PancreaticCancer immunotherapy
0
43
133
CellChat for systematic analysis of cell–cell communication from single-cell transcriptomics
1
69
302
We've been awarded an NIH R01 grant to further improve ShinyGO! 🎉 - Thank you for using it and citing our paper! - These support letters were essential! - This means the annotation databases will be updated for at least 5 years, also benefiting iDEP users. - Feel free to
New feature for GO enrichment analysis website ShinyGO v0.741 ( https://t.co/e9hHjIIhBT): Detailed genes info with links to NCBI and Ensembl. A fully customizable lollipop plot to visualize enrichment. Is it too fancy?
13
52
390
SpatialOne: end-to-end analysis of visium data at scale https://t.co/UStmFmVBOi 🧬🖥️ https://t.co/mPJIGM7UUr
2
16
44
Mapping cellular interactions from spatially resolved transcriptomics data https://t.co/9Ie1nIMmZ1 (read free: https://t.co/sFmeo6cOeK) Spacia software: https://t.co/tFoBvqdcpf 🧬🖥️
0
6
19
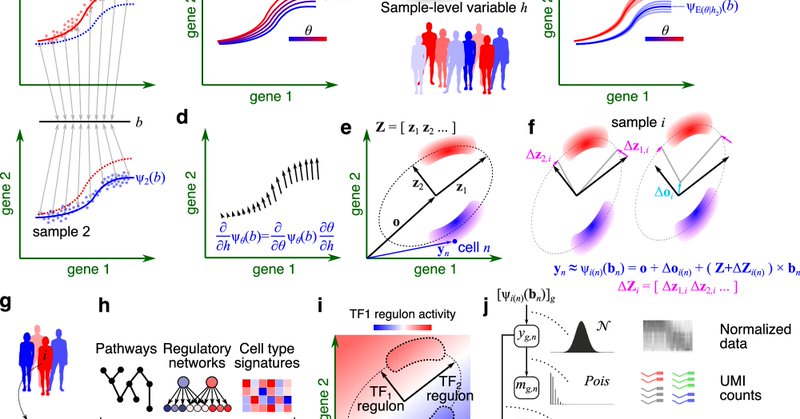
A unified model for interpretable latent embedding of multi-sample, multi-condition single-cell data
nature.com
Nature Communications - Single-cell analysis of multi-condition cohorts requires modelling the interaction between sample variables and cell states. Here, authors develop GEDI to enable...
0
8
43
Now out - WebAtlas pipeline for visualizing and processing integrated single cell and spatial transcriptomics data @naturemethods @bayraktar_lab @HaniffaLab @shazanfar @humancellatlas https://t.co/sSgxHwVl4Y
nature.com
Nature Methods - WebAtlas pipeline for integrated single-cell and spatial transcriptomic data
2
53
213
Many #SpatialTranscriptomics tools are out there! Which performs best? Which runs fastest? A nice resource here😆👇 16 clustering methods SpatialPCA https://t.co/UiMaUO7dr7 SpaGCN BANKSY GraphST STAGATE SEDR PRECAST conST DeepST CCST BASS ADEPT SpaceFlow BayesSpace ConGI 10
0
48
138
Benchmarking clustering, alignment, and integration methods for spatial transcriptomics👍 https://t.co/Y7unBn8LZW
link.springer.com
Genome Biology - Spatial transcriptomics (ST) is advancing our understanding of complex tissues and organisms. However, building a robust clustering algorithm to define spatially coherent regions...
0
21
73
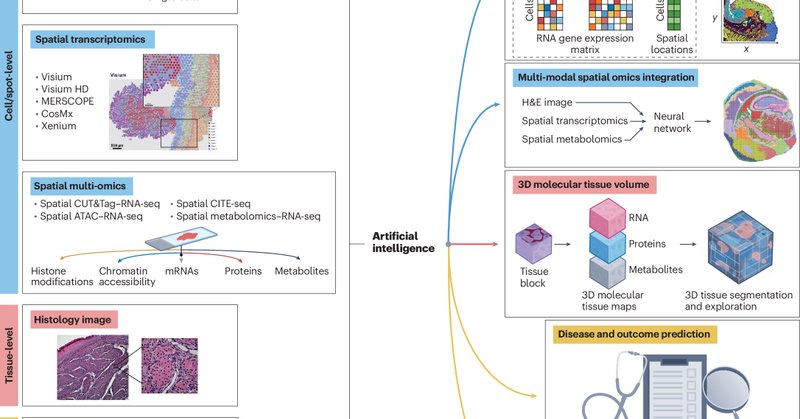
🤩Excited to share our @naturemethods comment on “Unlocking the Power of Spatial Omics with AI” https://t.co/i0ffuCEZUv co-authored with Kyle Coleman and Amelia Schroeder @UPennDBEI. Big thanks to @rita_strack for inviting us to contribute.
nature.com
Nature Methods - Spatial omics technologies have transformed biomedical research by offering detailed, spatially resolved molecular profiles that elucidate tissue structure and function at...
7
63
276