Yasuhiro Arimura
@ArmrYshr
Followers
904
Following
969
Media
64
Statuses
551
Elucidating the structural basis of biological events on chromosomes | chromatin, Xenopus egg extract, MagIC-cryo-EM | Assistant Professor at @HutchBasicSci
Seattle, WA
Joined September 2018
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
#Mステ
• 394828 Tweets
#Mステ
• 394828 Tweets
billie
• 285835 Tweets
CHASING THE SUN
• 131835 Tweets
Scottie
• 97954 Tweets
Valhalla
• 42825 Tweets
#のらりクラり呑もうの会
• 35886 Tweets
Louisville
• 31391 Tweets
ひーくん
• 28890 Tweets
ジュラシックワールド
• 16989 Tweets
トップバッター
• 15114 Tweets
しーちゃん
• 14549 Tweets
Mリーグ
• 12843 Tweets
Go City Go
• 12792 Tweets
ベビモン
• 11446 Tweets
NEW hair
• 10988 Tweets
佐々木朗希
• 10591 Tweets
西洸人くん
• 10402 Tweets
#大倉忠義オンライン飲み会
• 10120 Tweets
BACK TO THE BEGINNING
• 10006 Tweets
Last Seen Profiles
Pinned Tweet
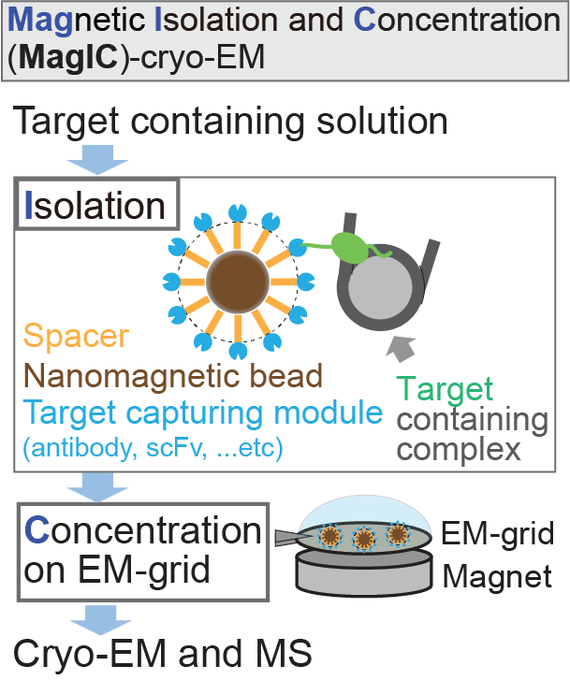
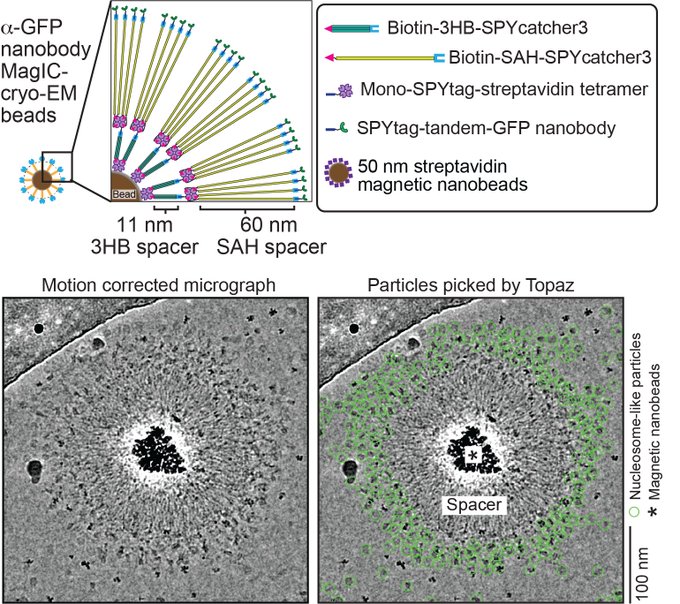
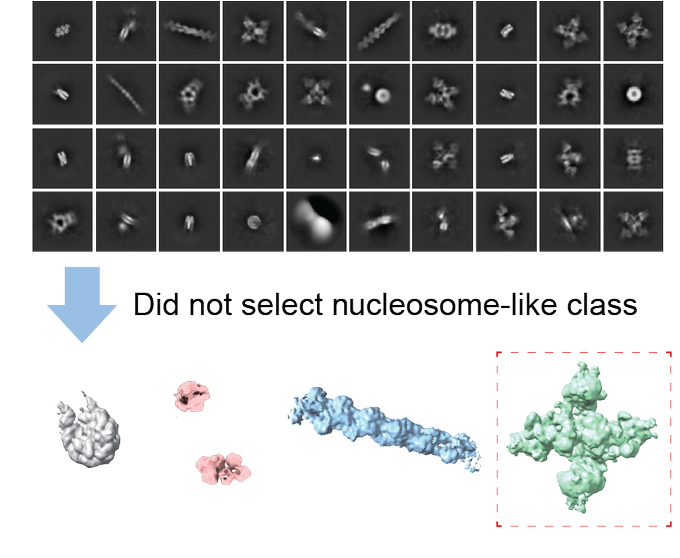
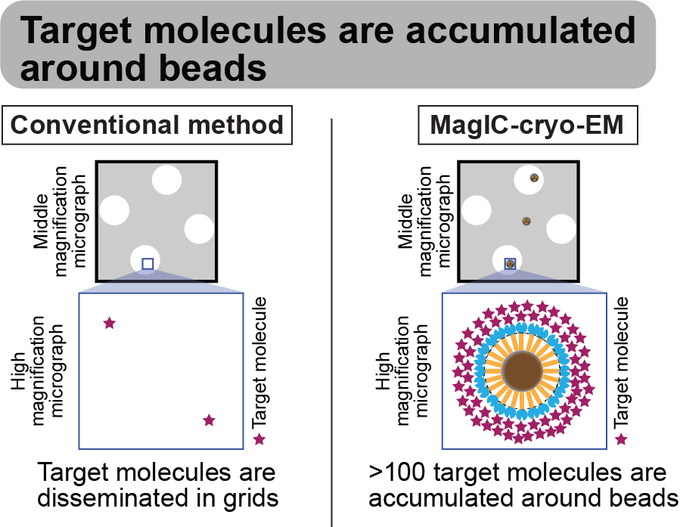
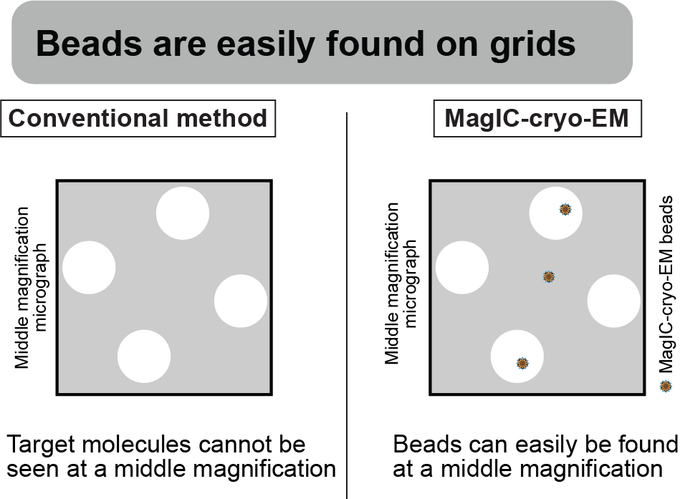
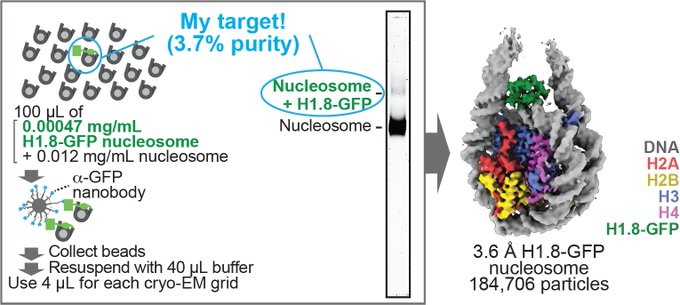
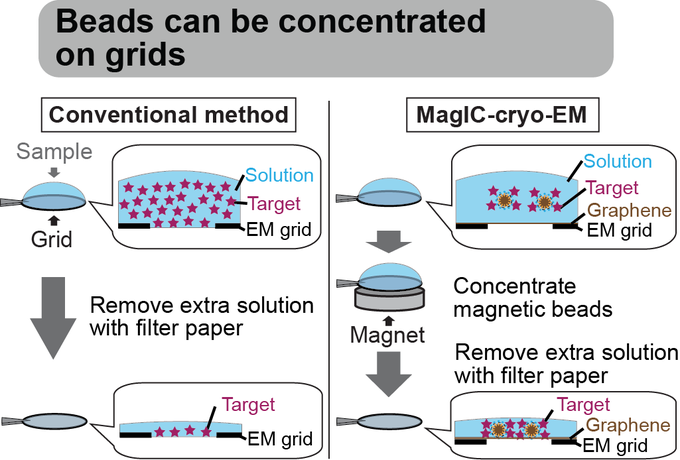
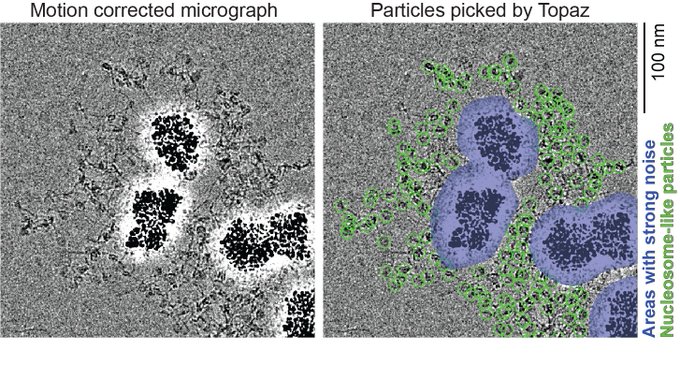
Magnetic Isolation and Concentration (MagIC)-cryo-EM is a new method to enable direct structural analysis of targets captured on nanomagnetic beads, which requires only 0.0005 mg/mL of targets in 100 µL crude solution to make 10 cryo-EM grids. I will explain the magic behind it!
9
118
430
Please repost!
Arimura Lab is hiring! We are seeking the next generation of MagICians (technicians and a postdoc) to elucidate the structural basis of events on chromosomes🧙♀️
No prior structural biology experience is required!!
Link
0
105
165
I am extremely excited to start my lab at Fred Hutch next year! Perfect environment to study chromatin! My lab aims to elucidate the structural basis of biological events on chromatin through the structural analyses of "native" chromatin complexes!
2023 was such an amazing year for the division, including the launch of the
@FredHutch
Postbac Program, the hiring of three of the brightest minds in their fields (
@C_P_Lapointe
,
@SRsrivatsan
,
@ArmrYshr
), and so much more. Read our 2023 Annual Report:
0
8
18
5
16
163
Plasmids for MagIC-cryo-EM are now available from Addgene! 🧙♀️
2
23
128
I am thrilled to begin my lab at my dream institute! 😆😆Recruitment ads will be posted in a couple of weeks. Stay tuned!
We are incredibly excited to welcome our newest faculty member,
@ArmrYshr
, on his first day in Basic Sciences
@FredHutch
! The Arimura Lab will study how chromatin functions, how it’s regulated, and what goes wrong in diseases like cancer. Learn more:
2
3
19
0
13
98
I am so excited to present in the Fragile nucleosome seminar!
Happy Friday everyone! Next week's seminar is going to be fantastic! We'll get to hear from two amazing structural biologists
@ArmrYshr
and
@lapassmore
!! **Recommended to attend live** Register here:
0
22
61
2
13
60
Check out the other new paper published today from
@KurumizakaLab
!
I've contributed to the first part of the work. Probably this one is my favorite Fig 1 among my works.
Everything started from a total failure experiment...
3
12
58
Stunning work by my Ph.D. mentor, Kurumizaka-sensei! He started RecA (bacterial homolog of Rad51) research during his Ph.D. work at the Shibata lab. Then, he started nucleosome research at the Wolffe lab. Now, these two finally unite in the cryo-EM structure! Congratulations!🎉
Our new paper about the structural mechanism by which RAD51 assembles at double-stranded DNA break sites on chromatin has been published in Nature!
#CryoEM
#Nucleosome
#RAD51
#DNArepair
11
121
514
0
7
56
Hey, chromatin enthusiasts in New York! I will present my data on the NYC chromatin club next Tuesday. See you at the meeting! (I am retweeting this to push me to prepare my presentation😅)
2
10
49
I'm thrilled to be part of the MCB program and help the next generation of talent thrive!
Join us in welcoming our newest faculty members, Dr. Yasuhiro Arimura (
@ArmrYshr
) from
@HutchBasicSci
and Dr. Devasena Ponnalagu (
@DPonnalaguLab
) from
@UWPharmacology
!
0
1
8
0
5
45
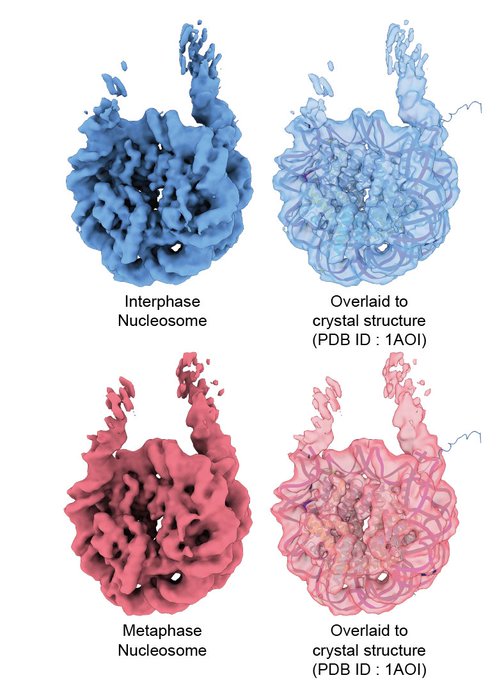
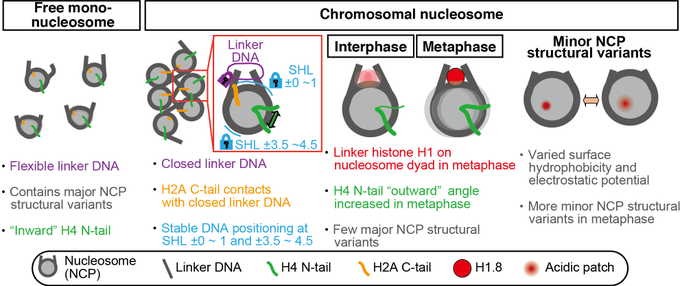
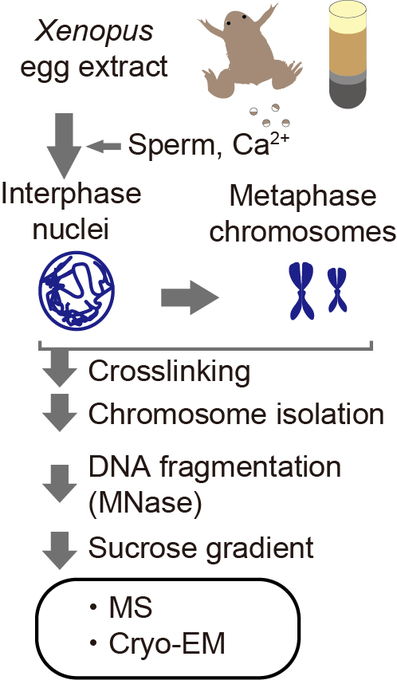
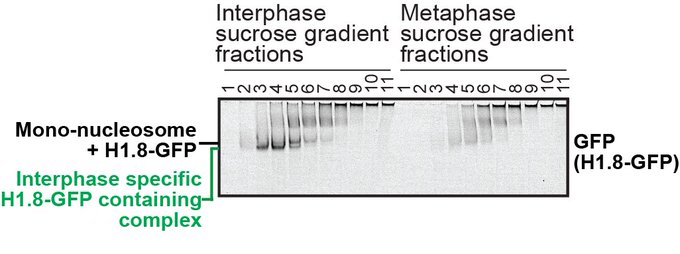
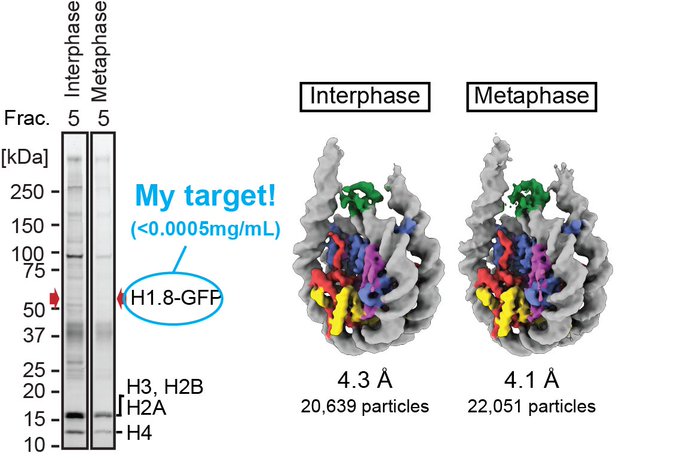
We determined the near-atomic resolution structure of nucleosome in interphase and metaphase chromosome by combining the Xenopus egg extract system I have learned at the
@HironoriFunabi1
and the nucleosome structural biology I have learned at the
@Kurimizakalab
.
1
5
31
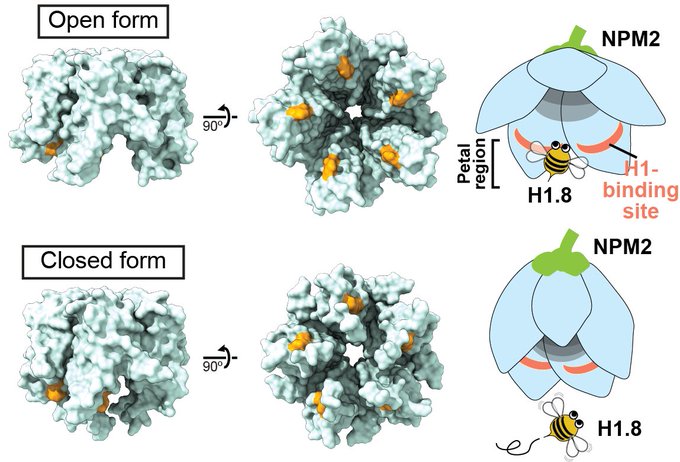
Check out the exciting paper from my fantastic colleague!! My latest preprint showed that the H1 tightly bound on the nucleosome dyad in the M phase. He has revealed the novel H1 function in the M phase: controlling the number of condensins and topo II on chromosomes!
0
7
30
I am on YouTube now😳
Just in time for the weekend! If you'd like to watch back Yasuhiro Arimura's talk on cryo-EM structures of nucleosomes in interphase and metaphase, you can find it on our YouTube channel! Thank you for sharing your work in our series,
@ArmrYshr
!
0
5
22
0
5
23
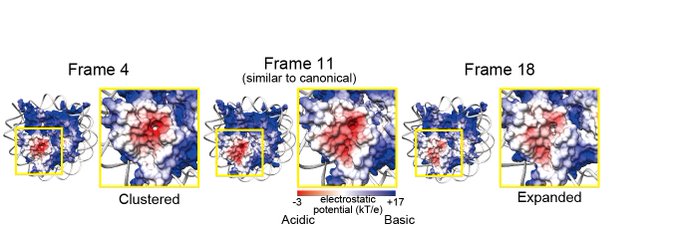
Cool!! ScFv for the acidic patch can somehow stabilize H1 on dyad (does it reduce NCP structural variation??). Also, I've never expected that H3 N-terminal goes this orientation in the chromatosome.
2
3
23
Arimura Lab is also participating in the Fred Hutch Postbaccalaureate Scholar Program! The scholar develop a foundation for a research career through chromatin biology research in our lab🧙🧙♂️🧙♀️
The Fred Hutch Postbaccalaureate Scholar Program is accepting applications! This program is for recent college grads who would benefit from research experience and training prior to applying to
#biomedical
PhD programs. Apply by Monday, April 1:
0
17
13
0
6
17
To improve my Twitter environment, I've made a bot to review English words/idioms I came across in PostDoc life in the US
@StudyingYasu
Feel free to follow it if you are also an English learner🤓
0
2
16
I previously found that the CENP-A nucleosome forms outward H4 tail confirmation and this confirmation somehow facilitates H4K20me1 modification by SET8(AKA PR-SET7)
This work explained why. H4 tail orientation in CENP-A nucleosome is ready to bind SET8!!
1
3
14
Check out my colleague's Tweetorial (Xtorial?) about her amazing finding: the identification of the new hemimethylated CpG binding protein!
Very excited to announce our pre-print! Together with the labs of
@NishiyamaAtsuya
and
@KyoheiArita
, we’ve discovered that the protein CDCA7 (a long-time favourite in the Funabiki lab) exhibits a striking ability to sense hemi-methylated DNA. 1/7
4
9
41
0
0
11
Virus nucleosome week!🤯🤯
Indeed, some viruses encode histone doublets!
#viralnucleosomes
are fragile, and have extended linker DNA. Check out the structures. AND they are specifically recruited to the Marseillevirus factory and virus, shown by our collaborators
@chantal_abergel
.
.
6
32
159
0
0
10
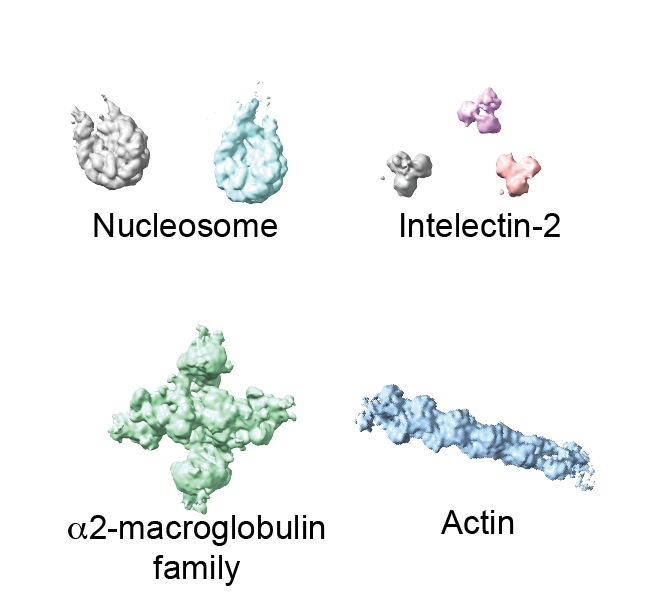
With tremendous help from Seth Darst lab and
@RubyFroom
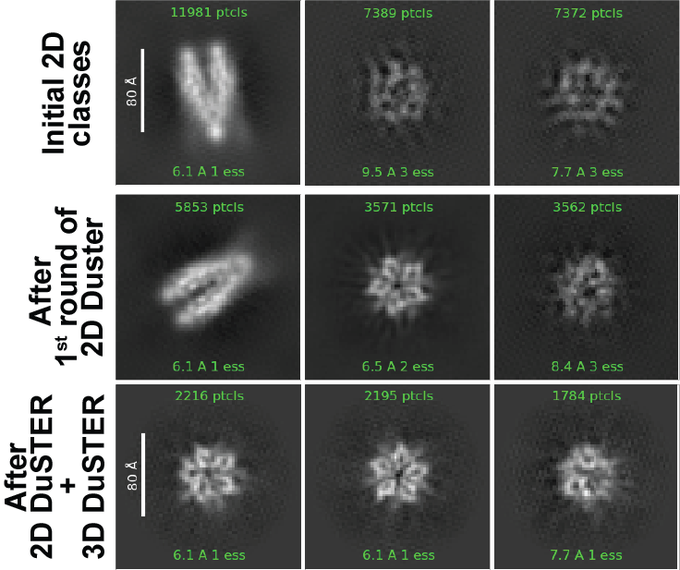
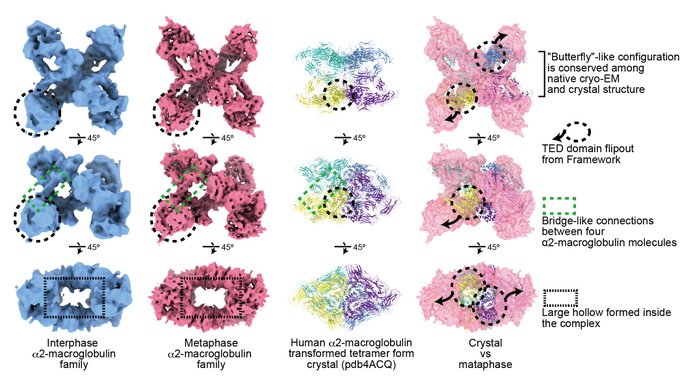
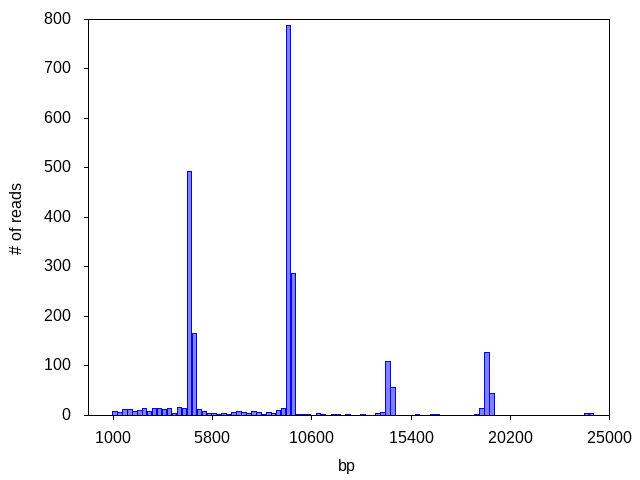
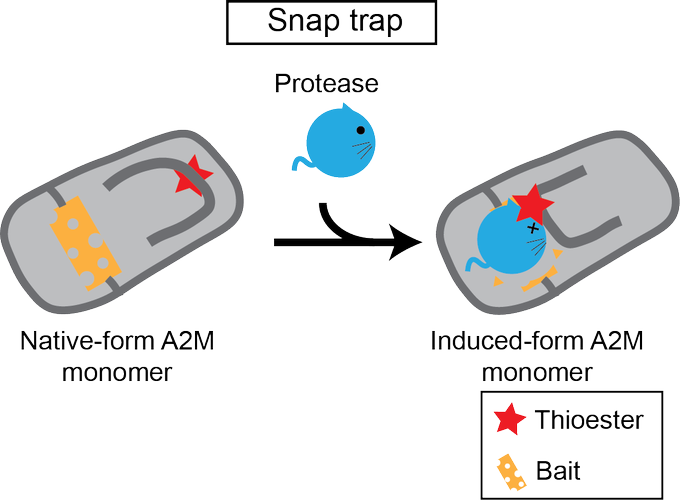
in his lab, we determined the structures of the five most abundant complexes (nucleosome, chromatosome, alpha2-macroglobulin, intelectin-2, actin filament) at once in an unbiased way (Fig. 2).
1
0
10
I used this 3DVA in my latest preprint. The 3DVA (& cryoSPARC of course) is amazing🤯 Thank you for developing this awesome method!!
Interested in continuous heterogeneity+flexibility in
#cryoEM
? Check out our updated
#cryoSPARC
3D Variability Analysis paper on
@biorxiv
!
It’s great to see 3DVA being used in the wild to resolve beautiful proteins in motion 😀
1
14
57
1
3
9
Congratulations, Daniel🥳
The cryo-ET structure of the native kinetochore is more flexible and larger than anticipated!
(Also, MagIC-cryo-EM contributed in an unanticipated way: makes proper thickness ice)
The bulk of my grad work is finally out in the world! Big thanks to everyone in
@SueBiggins
lab and collaborators including
@HHMIJanelia
,
@WijeratneS
,
@ArmrYshr
,
@HironoriFunabi1
, and others not on this site. Wouldn't have been possible without great collaborators!
1
11
30
0
0
8
A really cool work from Seth Darst's lab (one of my cryo-EM mentors)!! Congratulations🎊
Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy
#bioRxiv
0
1
5
0
0
7
Christian is an amazingly kind man with a broad range of profound knowledge. I do recommend this position!
Post-doc position available in my lab for a collaborative project together with Pascal Meier
@MeierLabICR
and Jon Pines
@PinesLab
to study innate immune responses to radiation. Apply at or message me for more information.
4
48
68
1
2
8
Here are the links for my Tweetorial!! I hope you enjoy reading them!
MagIC:
1
2
6
Thank you for many comments to our preprint!! We modified the title.
Thanks for overwhelmingly positive responses to our recent preprint by
@ArmrYshr
!
>1200 downloads!
In response to a comment by
@RadoDanev
, we uploaded a V2 with a modified title.
Nucleosome structural variations in interphase and metaphase chromosomes
1
1
46
0
0
6
Wow😍
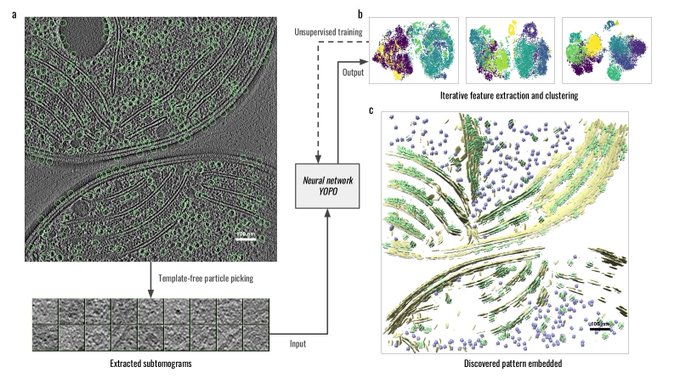
Welcome check out our new manuscript. DISCA: high-throughput cryo-ET structural pattern mining by deep unsupervised clustering.
The work was led by my PhD student Xiangrui, in collaboration with
@yiweichang
and Mahamid teams
@Liang_Xue_
.
1
26
93
0
0
6
I thank my super supportive mentor
@HironoriFunabi1
, who kindly learned about A2M from scratch with me with turning himself into a Dracula.
.
26/n
1
1
5
🤯
Please see our Cryo-EM structure of nucleosome-like particles formed by Marseillevirus histone fusions and DNA online in
@NatureSMB
. Fantastic work by
@Marco_Va1
and
@Xagbomson
! Special thanks to our collaborators! Here is a link:
15
69
273
0
0
5
@HironoriFunabi1
Thank you so much for your support! The five years I spent in the Funabiki lab were amazing. Combining your expertise with mine, we developed unique approaches. You also connected me with so many scientists. Thank you so much!
1
0
5
Molecular replacement of
crystallization contaminants👀
AlphaFold Protein Structure Database for Sequence-Independent Molecular Replacement
#bioRxiv
0
18
81
0
0
5
Amazing!!🤯
We’re excited to share cryo-TomoSim (aka CTS), our
#cryoET
modeling and simulation software, in addition to our new preprint documenting its use in training regressive denoising and semantic segmentation U-Nets.
7
72
334
1
0
5
Also, this work was only possible because I was trained by the nucleosome enthusiast, Kurumizaka-sensei
@KurumizakaLab
during my Ph.D. training.
1
0
4
or the youtube video at Fragile Nucleosome Seminar Series!
Just in time for the weekend! If you'd like to watch back Yasuhiro Arimura's talk on cryo-EM structures of nucleosomes in interphase and metaphase, you can find it on our YouTube channel! Thank you for sharing your work in our series,
@ArmrYshr
!
0
5
22
1
1
3
@SaKanYoo1
通りすがりです。プラスミド同士が複数くっつくことは、昔ながらの方法で検出できないだけで、実はよく
あるみたいです。 ただ、例に挙げられた二つの制限酵素は、巨大な回文配列になってしまうので、大腸菌の中で生き残らないのではないでしょうか。
I think
@plasmidsaurus
is just trolling me 💀"Propagate a plasmid in MG1655, what's the worst that can happen?"
5
3
40
1
1
3
We showed the mechanism that enables the H4K20me1 accumulation on CENP-A nucleosome by X-ray crystallography, biochemistry (collaborate with Kimura Lab
@Cell_Tokyo_Tech
), and Quantitative ChIP-Seq (collaborate with Fukagawa Lab
@tatsuofukagawa1
).
1
0
3
@RubyFroom
@HironoriFunabi1
Thank you so much for your advice and coaching on data processing!! The analyses in the paper were only possible because of your help!!
0
0
3
@LugerLab
@UCSFChimeraX
@biorxivpreprint
Wait.. is it a split nucleosome?? I can't wait to read the paper
0
0
3
@LugerLab
@DShechter
Thank you for your interest! Here is a link for signing up for the NYC chromatin club mailing list to get the zoom link (Please hit "subscribe" on the top left). I assume the club is open for worldwide chromatin people since the club tweet this public 🤔
3
0
3
In our previous work, we determined the structures of the nucleosomes and H1-bound nucleosomes formed in chromosomes. However, >300 other chromatin-associated proteins detected by MS remained unsolved.
1
0
3