Lori Passmore
@lapassmore
Followers
5K
Following
2K
Media
117
Statuses
1K
Molecular biologist. Group Leader @MRC_LMB. Fellow @clarehall_cam. Optimist. (she/her) Views/opinions are my own.
Cambridge UK
Joined January 2015
I am no longer actively using this account. Please find me on the other place. 🦋
0
1
14
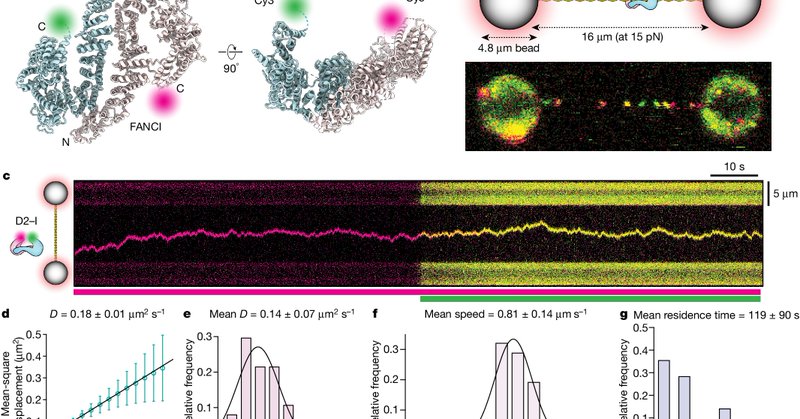
We have an open #postdoc position to study the molecular mechanisms of #DNArepair !🧬🔬 The project will build on recent findings https://t.co/TDxCrkgZKb and established collaborations, using #cryoEM and #SingleMolecule imaging. Access to fantastic facilities @MRC_LMB .
nature.com
Nature - FANCD2–FANCI is a sliding clamp that diffuses on double-stranded DNA but stalls when it reaches a single-stranded gap, providing a unified molecular mechanism that reconciles the...
2
29
60
CryoEM community: we encourage you to submit applications to the Almeida Award #cryoEM BPS subgroup:
Mark your calendars for the BPS CryoEM Subgroup Symposium, on Subgroup Saturday at @BiophysicalSoc Annual Meeting 2025! Also, we have a new award, the Almeida Award, for a mid/senior level woman who has made outstanding contributions to #cryoem. See you there!
0
8
15
Unfortunately I had to leave the conference very suddenly and unexpectedly due to a family emergency. I really missed meeting and speaking with so many people. Many apologies - I would be happy to zoom meet with anyone who wants to chat - please drop me an email!
0
0
0
I presented this work at #EESRNA @embl
@EMBLHeidelberg An amazing meeting organised by @LabConti @OlivierDuss @torbenheick and Karla Neugebauer!!
New work from the lab! James Stowell led this project showing the importance of multivalency and phospho-regulation in mRNA decay. This has many parallels with other mechanisms in gene expression... 🧵 1/ #RNA #NMR #bioRxiv @MRC_LMB @ERC_Research
1
1
34
The enzymatic machineries of gene expression (deadenylation complexes, RNA polymerases) are highly conserved. .... but the evolutionary plasticity of IDRs found in a multitude of regulators (RNA adaptors, transcription factors) allows rewiring of gene expression programs. 10/
1
0
3
This interaction mechanism has parallels with other processes in gene expression including transcription and decapping. Continuous tuning likely occurs within multiple steps of gene expression ... 9/
1
0
1
We conclude that phosphorylation can result in a rheostat-like response on the multivalent interaction allowing ‘tuning’ of deadenylation activity. This is a common principle across multiple RNA adaptors (e.g. Pumilio proteins, TTP/ZFP36) in yeast and human complexes. 8/
1
0
1
Excitingly, this multivalency can be regulated by multi-site phosphorylation of the IDR. This modulates IDR charge and dynamics to ‘tune’ deadenylation rate. 7/
1
0
1
Targeted mutations in individual residues across the IDR (or in dispersed surface pockets of Ccr4-Not) results in a *graded* decrease in deadenylation activity, pointing towards a multivalent interaction. 6/
1
0
1
In this work, James used NMR with @connyyu , crosslinking mass spec with @RappsilberLab and biochemical reconstitution to discover that dispersed regions within the IDRs interact with multiple conserved binding sites on Ccr4-Not. 5/
1
0
5
Intrinsically-disordered regions (IDRs) in RNA adaptors interact with Ccr4-Not. This promotes deadenylation of bound transcripts. 4/
1
0
4
But how are specific mRNAs targeted for decay at the correct time? Many different ‘RNA adaptor’ proteins (RNA-binding proteins) tether specific transcripts directly to Ccr4-Not. 3/
1
0
2
Poly(A) tails control translation and stability of eukaryotic mRNAs. The Ccr4-Not deadenylation complex shortens these tails to control gene expression in a regulated and context-dependent manner. 2/
1
0
1
New work from the lab! James Stowell led this project showing the importance of multivalency and phospho-regulation in mRNA decay. This has many parallels with other mechanisms in gene expression... 🧵 1/ #RNA #NMR #bioRxiv @MRC_LMB @ERC_Research
7
42
186
Great fun with the @RappsilberLab ! Photo-crosslinking improves the contrast of crosslinking mass spectrometry.
New preprint! We show that photo-crosslinking dramatically improves structural insights into protein complexes compared to traditional crosslinking MS methods. https://t.co/sKlI9KTtRh
0
1
3
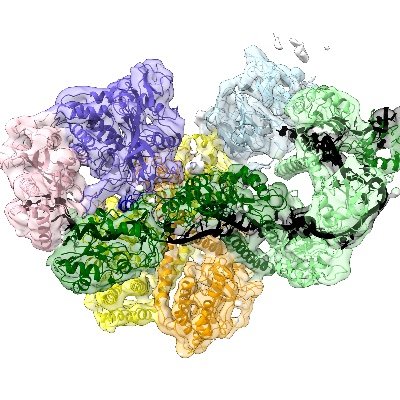
Very cool structure!
Excited to share our latest research on co-translational mRNA decay by the exosome in human cells! We uncover how the exosome-SKI complex is recruited to ribosomes and propose a novel role for SKI in recognizing collided disomes. @MPI_Biochem 👇👇👇
0
0
15
I am really looking foward to working with @SjorsScheres as Joint Head of Structural Studies @MRC_LMB !
11
1
111