William Allen
@weallen1
Followers
2K
Following
458
Media
18
Statuses
183
Biology of homeostasis & resilience. New tools to map + control cells. Asst prof @StanfordDevBio. @Harvard Junior Fellow, neuro Ph.D. @Stanford, MPhil @MRC_LMB
Stanford, CA
Joined June 2008
Very excited to announce that I'll be starting a laboratory studying the biology of homeostasis as an assistant professor in the Department of Developmental Biology (@DevBioStanford) at @Stanford in July! https://t.co/KjDvPUa4LI
26
28
309
My paper is out!! 🎉 "Opponent control of reinforcement by striatal dopamine and serotonin", @Nature Here, we show that dopamine and serotonin signals form a gas-brake system for reward in the mammalian brain 🧠 THREAD ⬇️ 1/n
40
169
888
Delighted to share this preprint from Prof. Rong Fan Lab @RongFan8 and Prof. Sidi Chen lab @sidichen on Spatially Resolved in vivo CRISPR Screen Sequencing via Perturb-DBiT. Congatulations, Alev, Xiaolong, Feifei, Paul, Sidi and Rong!
Thrilled to share Perturb-DBiT — spatial unbiased in vivo perturb-seq with genome-scale CRISPR libraries! Hope you enjoy reading it over the Thanksgiving week 🥰 Kudos to @AlevBaysoy, Xiaolong, Feifei, and Paul. & collaboration with @sidichen, Hongbo Chi. https://t.co/IcUPAG5k8P
1
3
23
Thrilled to share Perturb-DBiT — spatial unbiased in vivo perturb-seq with genome-scale CRISPR libraries! Hope you enjoy reading it over the Thanksgiving week 🥰 Kudos to @AlevBaysoy, Xiaolong, Feifei, and Paul. & collaboration with @sidichen, Hongbo Chi. https://t.co/IcUPAG5k8P
biorxiv.org
Perturb-seq enabled the profiling of transcriptional effects of genetic perturbations in single cells but lacks the ability to examine the impact on tissue environments. We present Perturb-DBiT for...
5
61
281
delighted to share this exciting news with Prof. Xiaowei Zhuang @zhuanglab and Prof. Jonathan Weissman @JswLab, on their latest preprint reporting Perturb-Multi: a multimodal in vivo pooled CRISPR screen 🤩 congrats Reuben and William @weallen1 👏🍾 https://t.co/BMvXVDCVX8
1
3
17
Excited that the Zhuang Lab is now on X!
Check out our preprint on Perturb-multi ( https://t.co/lFeRYA0lco), a multi-year collaborative effort that enabled multi-modal in vivo genotype-phenotype mapping by pooled genetic perturbations and high-dimensionality phenotype readout by imaging and sequencing.
0
1
9
Check out our preprint on Perturb-multi ( https://t.co/lFeRYA0lco), a multi-year collaborative effort that enabled multi-modal in vivo genotype-phenotype mapping by pooled genetic perturbations and high-dimensionality phenotype readout by imaging and sequencing.
In collaboration with Reuben Saunders, @JswLab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! https://t.co/iJ8hi3ddz4
1
9
37
New work from @arcinstitute on the cover of Science this week!
A new Science study presents “Evo”—a machine learning model capable of decoding and designing DNA, RNA, and protein sequences, from molecular to genome scale, with unparalleled accuracy. Evo’s ability to predict, generate, and engineer entire genomic sequences could change the
10
26
308
Check out the preprint describing Perturb-Multi from Arc Ignite Awardee @weallen1, Reuben Saunders (now at Arc), @JswLab and Xiaowei Zhuang. A general and scalable approach applied here to mouse liver, Pertub-Multi generates rich, multimodal data that can generate fundamentally
In collaboration with Reuben Saunders, @JswLab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! https://t.co/iJ8hi3ddz4
1
5
32
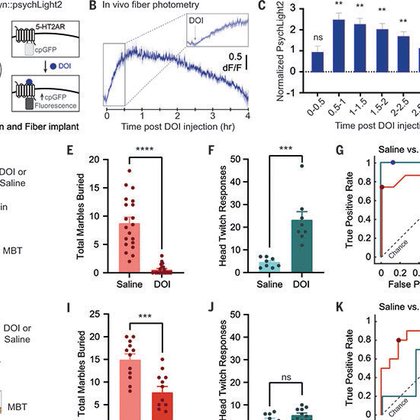
Happy to share the lab’s latest research tagging psychedelic-activated neurons in mice. See the post below from first author @jessie_muir for a tweeprint on the work. Thanks to wonderful collaborators @DEOlsonLab @LinTianPhD and to our reviewers/editor! https://t.co/Gt6iGbLWdn
science.org
Psychedelics hold promise as alternate treatments for neuropsychiatric disorders. However, the neural mechanisms by which they drive adaptive behavioral effects remain unclear. We isolated the...
Excited to share the Kim Lab’s newest paper in @ScienceMagazine with Sophia Lin (twitterless) and @tinakim_neuro ! We look at circuit mechanisms of psychedelic action underlying therapeutic-like effects in the PFC. https://t.co/zqq96eLRHB
17
29
168
One key component of Perturb-Multi is in vivo Perturb-seq of formaldehyde-fixed tissue, enabled by the Flex reagents from @10xGenomics.
In collaboration with Reuben Saunders, @JswLab, and Xiaowei Zhuang, we are very excited to release Perturb-Multi: a platform for pooled multimodal genetic screens in intact mammalian tissue. Check it out! https://t.co/iJ8hi3ddz4
1
11
66
My lab at @DevBioStanford ( https://t.co/sqIRzJcZQA) is looking for motivated postdocs and graduate students who are excited about developing new technology and applying in vivo genetic screens to reverse-engineer tissue development, homeostasis, and degeneration – contact me at
allenlabstanford.org
0
2
12
This was an amazing team effort over multiple years, bringing together diverse expertise from @Harvard @HarvardCCB @MCB_Twitter and @WhiteheadInst @MIT @HHMINEWS. We look forward to expanding to the genome-wide scale and extending to other organs!
1
0
6
Along with recent spatial developments – e.g. Perturb-FISH, Perturb-View (@eric_lubeck) and CRISPR-Map (@GaublommeL) – and fantastic in vivo work from @XinJin and @randall_platt, this is an extremely exciting time for pooled screening, particularly with large-scale AI efforts
1
1
9
This work builds on foundational Perturb-Seq dating back to 2016 from the Weissman @JswLab and Regev Labs, and pioneering work on imaging-based pooled genetic screening using MERFISH dating back to 2017 from the Zhuang and Elf labs, and using ISS from 2019 from @BlaineyLab.
1
0
5
By integrating sequencing and imaging, we can achieve insights that would be difficult from each modality alone, such as convergent effects of mechanistically distinct pathways that all produce cellular steatosis.
1
0
4
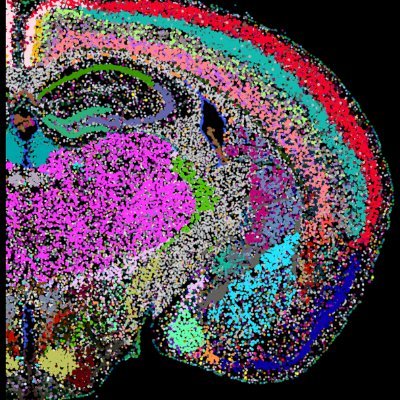
A core aspect of liver organization and function is hepatocyte zonation. Our data revealed both expected regulators of Zonation (Wnt signaling, O2 sensing) as well as potentially novel pathways, in a cell autonomous manner. This work reveals the highly dynamic nature of zonation
1
0
4
Because these genetic perturbations are introduced into a living mouse, we can look at the interactions between gene function and physiological state. E.g., we can identify functional re-wiring following overnight fasting, which leads to dramatic metabolic state-dependent
1
0
4
We can then zoom in to identify the specific functional effects of perturbing different genes in an automated and unsupervised way. For example, combining subcellular imaging with deep learning-based embeddings identified Npc1 as a regulator of lysosome structure (measured
1
0
4
This rich, multimodal data enables multiple analyses of how genetic perturbations affect multiple aspects of tissue structure and function from the same animal. At a high level, global multimodal analysis of different perturbations allows us to group them into shared pathways.
1
0
6
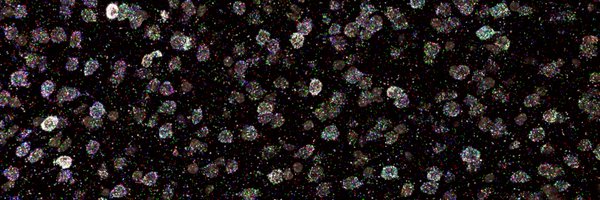
In mosaic perturbed mouse livers, we detected small clumps of cells with the same perturbation and measured potent transcriptional phenotypes.
1
0
6