Pranam Chatterjee
@pranamanam
Followers
2,611
Following
336
Media
34
Statuses
479
Designing proteins to program biology! 🧬💻🧫 Assistant Professor at @DukeU | Co-Founder @GametoGen and @UbiquiTxINC | Formerly @MIT '16 '18 '20 and @harvardmed
Cambridge, MA
Joined August 2015
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
#JISOOxDiorSS25
• 225710 Tweets
ブロック
• 214158 Tweets
APO WITH DIOR IN PFW
• 147552 Tweets
刀剣男士
• 70888 Tweets
#ข้ามฟ้าเคียงเธอQ6
• 62481 Tweets
The Next Prince Q6
• 59593 Tweets
イーロン
• 41397 Tweets
#バナナサンド
• 40771 Tweets
オリックス
• 35991 Tweets
イナズマイレブン
• 19553 Tweets
T-岡田
• 18675 Tweets
引退試合
• 18089 Tweets
TAYNEW x GRABFOOD LIVE
• 17396 Tweets
#仰天ニュース
• 12472 Tweets
初代リメイク
• 10905 Tweets
Last Seen Profiles
Hi everyone! Excited to share that I will join the amazing faculty at
@DukeU
as an Assistant Professor of Biomedical Engineering in July! We will design therapeutic proteins for genome editing, proteome editing, and ovarian cell engineering! 💻→🧬→🧫→🏥
25
7
170
Super excited to share our new protein language model: FusOn-pLM, developed by my wonderful first-year PhD student
@SophieVincoff
and her team of amazing undergrads!! 🎉
Sure, we know AlphaFold3/RFDiffusion and other structure prediction/design methods are great -- but what
8
36
171
#NeurIPS2023
was so much fun! My favorite talk from the workshop has to be from
@PrescientDesign
! Similar to
@OpenAI
's Consistency Model formalism, they sample a score-based data manifold with one step denoising for antibody design!
#GenerativeAI
3
21
138
An exciting
@biorxivpreprint
update on our recent PepPrCLIP model!🌶️📎 Hopefully, you all remember PepPrCLIP, where we apply Gaussian perturbations to the peptidic latent space of ESM-2 to de novo generate naturalistic peptides, and then input these new peptide sequences into a
0
21
128
Okay, this is SO COOL from
@Mila_Quebec
: Aaren, which leverages a parallel prefix scan algorithm, can be trained in parallel (Transformers) but only requires constant memory (RNNs)! 🤩 It's kind of similar to RWKV and Linear Attention, but unlike those methods, Aaren exactly
0
20
111
Excited to share our newest Cas9 out in
@NatureComms
! 🥳 We recombine our Sc++ enzyme (NNG) with the PID of
@BKleinstiver
's SpRY (NRN) to generate a PAM-flexible chimeric Cas9 = SpRYc! 🌶️ SpRYc can edit at diverse NNN loci -- take it out for a spin! 🧫
3
27
110
Exciting news from the lab! 👀 We have updated PepMLM on the
@arxiv
with new computational and experimental benchmarking results! 🥳
Just as a reminder: we trained PepMLM via a novel span masking strategy that positions peptide binder sequences at the C-terminus of their target
5
22
99
🧵 (1/4) As a brand-new PI, I'm psyched to present my group's first preprint! Here, we apply
@OpenAI
's CLIP architecture to design target-specific peptides, fuse them to E3 ubiquitin ligases, and degrade pathogenic proteins in a pipeline called Cut&CLIP. ✂️
7
11
92
Just getting around to posting about this beautiful paper from
@DreamFoldAI
!! Learning a joint representation based on SOTA sequence/structure embeddings and decoding the joint representations of the inputs into SE(3) vector fields is very clever -- my hunch is that the ESM-2
We are thrilled to announce FoldFlow-2: our new SOTA protein structure generative model.
w/
@Guillaume_hu
James V
@FatrasKilian
Eric TL
@PabloLemosP
@riashatislam
@ChengHaoLiu1
@jarridrb
@tara_aksa
@mmbronstein
@AlexanderTong7
@bose_joey
🪄 Blog post:
3
24
107
1
18
88
Today, I "officially" begin my faculty position at
@DukeU
! 🥳 Super excited to welcome my wonderful team of postdocs, PhD/Masters students, and undergrads from around the globe to our little but mighty lab in Durham ― let's change the world together! 💪🏾
5
7
78
SaLT&PepPr is published in
@CommsBio
! 🥳 Here, we fine-tune the ESM-2 pLM to identify peptidic binding sites on target-interacting partner sequences. We fuse these "guide" peptides to E3 ubiquitin ligases to degrade disease-causing proteins! 💻➡️🧫 (1/n)
5
15
77
How can AI help us cure rare diseases? In the Aug/Sep
@WorkingAtDuke
magazine, I talk about my lab
@DukeUBME
and how we’re training generative language models to help us design drugs to target complex proteins. Keep an eye out for the magazine in your mailbox!
@dukeresearch
1
8
72
Such a beautiful study from our friends in
@rohitsingh8080
’s lab here at
@DukeU
! An autoencoder on top of ESM-2 for fixed dimensionality is brilliant, and perfectly suited for this problem, one that has plagued delivery and other drug design applications. 🌟 I can imagine so many
2
16
70
Hi everyone! I am very excited to announce that our broad, efficient, and specific enzyme, Sc++, has been published in
@NatureBiotech
! 🤩 An incredible collaboration with
@njakimo
@JooyoungLee10
@SontheimerLab
@medialab
@UMassMedRTI
🎉🎊 Check it out! 🔽 (1/5)
3
16
63
This is such cool work published in
@NaturePhysics
! 🤩 An N-terminal unstructured and flexible glycine–serine peptide tag can accelerate nuclear import rates for stiff protein cargos by increasing their deformability. Definitely useful for my experimentalists to deliver our
0
4
60
Well here it is: ESM3!! 🌟 The training regime is a bit different than your classic ESM-2/BERT model: you can start with a fully masked sequence and iteratively unmask. Super similar to our PepMLM model with ESM-2, which showed significant promise at full masking and unmasking.
1
9
56
I'm so excited to co-host the
@gembioworkshop
at
#ICLR2024
in Vienna!! 🥳To both my computational and experimental friends, please reach out if you are interested in submitting a paper or serving as a reviewer -- it should be an incredible, one-of-a-kind meeting! 🌟
Announcing the Generative and Experimental Perspectives for Biomolecular Design workshop at
#iclr2024
!
We hope to bring together researchers in ML and experimental biology to accelerate progress on real-world applications.
Website:
Paper deadline: Feb 3
1
52
132
3
11
53
A little late, but super excited to share a great collaborative work by our team at Duke, my postdoctoral lab of
@geochurch
, and my company
@GametoGen
to engineer a functioning human ovarian follicle from iPSCs using our STAMPScreen method! 🧫
1
7
50
After almost three years of painstaking effort, today, I'm excited to present my team's amazing work on specifying human oogonia via combinatorial transcription factor induction in iPSCs! 🥳 An incredible collaboration with my good friend
@krammecc
!
2
4
44
Beautiful stuff
@AlexanderTong7
@mmbronstein
and team!! MFM enables generative models to accurately match vector fields on data manifolds -- so many cool applications: LiDAR navigation, unpaired image translation, single-cell RNA trajectory inference...such impressive work! 😊
If data lives on a manifold, how do we design meaningful interpolations between marginals? We present Metric Flow Matching (MFM)…
@PPotaptchik
@TeoReu
@leoeleoleo1
@AlexanderTong7
@mmbronstein
@bose_joey
@Francesco_dgv
🔗Dive in here:
🧵 (1/12)
7
63
342
1
11
43
Excited that my first company
@GametoGen
has just raised our Series B!!🧫 This is the amazing work of our incredible CEO
@DinaRadenkovic
and CSO
@krammecc
who have led the company to new heights since
@DinaRadenkovic
,
@martinvars
,
@krammecc
, and I decided to get this off the
The AlleyWatch Startup Daily Funding Report for 5/29 ft:
STARTUPS:
@GametoGen
@authcaptives
FOUNDERS:
@dinaradenkovic
@martinvars
@pranamanam
@authcaptives
LEAD INVESTORS:
@twosigmavc
@firstmarkcap
#NYCtech
#startup
#funding
#VC
0
1
3
4
3
41
A very cool use of graph-based diffusion for enzyme evolution! Also love the application to pAgo, which can be a very powerful programmable gene editing tool with improvements.
#GenerativeAI
#ProteinDesign
0
9
41
Happy to share our early work on generating binding peptides conditioned ONLY on the target sequence! 🌟PepMLM masks cognate peptides at the end of target protein sequences, and tasks ESM-2 to fully reconstruct the binder region. 😷
#GenerativeAI
1
4
38
Training a VAE-based representation model, performing diffusion on the latents to generate new aptamers, with SPR results show an average 3-fold Kd reduction? Probably needs a bit of refinement and validation, but so cool! 🫨
#GenBio
0
4
38
Hi all! We have released a preprint on our novel SaLT&PepPr language model to design peptide-guided degraders using PPI information. 🥳 An incredible effort from
@garykbrixi
and team at
@DukeU
, and our collaborators at
@Cornell
with
@mattdelisa
! 🤜🏾🤛🏾
1
6
36
Today is my happiest day as a scientist: my incredible undergrad
@SabrinaKoseki
was awarded the
@NSF
GRFP! 🍾 I've mentored Sabrina for 3+ years now, and her brilliance, curiosity, and diligence EPITOMIZES scientific excellence. I've been so lucky to have her as my student!😊
3
2
35
Super excited to announce the publication of our "AA" targeting Cas9, iSpyMac, in
@NatureComms
! 😍 Another incredible collaboration with
@njakimo
@JooyoungLee10
@SontheimerLab
@medialab
@UMassMedRTI
🎊Check it out 🔽 (1/5)
6
6
32
Nice! A great application of Mamba on concatenated, homologous protein sequences! 👍 Happy the authors mentioned our recent PTM-Mamba model, where we showed the application of Mamba to represent modified sequences! Overall, SSMs seem to have a found nice home in biology! 🧬
2
5
32
The Programmable Biology Lab is all set up at
@DukeU
— all in 1.5 days! 💪🏽 Shoutout to my amazing Harvard CS undergrads, Garyk and Suhaas, for helping make this happen. 🌟Time to design some programmable proteins! 💻➡️🧫
5
0
31
We're so grateful to
#EndAxD
for believing in our binder-guided degrader technology, powered by
#GenAI
, to study and treat Alexander disease. 💻🧫 Thank you to
@thomas_wagner
and little Max for inspiring my lab to study, treat, and cure AxD! 🙏🏾
5
3
29
Check out our lab's perspective on generative design of therapeutic binders published in COBME: ! 💻🧫 Great work from my PhD students:
@LeoChan213
@lauren_hong11
@vivi_64_
@SophieVincoff
Feel free to cite the review in your next manuscript! 😊
#GenerativeAI
1
7
27
Remember
@DeepMind
's protein folding algorithm, AlphaFold2? I'm excited to share our team's early work on our own protein structure prediction pipeline! (1/3)
1
7
26
My last day together at the
@medialab
with my superhero team of undergrads,
@SabrinaKoseki
, Teodora, and Emma! So very proud to have watched you three become such brilliant young scientists, and so privileged to have mentored you as my first students! 🌟🥹
0
1
25
As the second week of my lab's existence comes to a close, I'm super grateful for the dedication and commitment of my students to our (quite lofty) goals. Yeah, we may not be the most well-known bunch, but we're about to do big things! 💪🏾
#FearlessFriday
1
0
24
An integrated pLM trained on BOTH sequence and structure! 🤩 We're excited to leverage these embeddings in our lab -- the T5 architecture is super powerful for design! So many congrats to
@HeinzingerM
and the
@rostlab
for another pioneering work! 👏
1
2
24
Hi friends! Just a gentle reminder that our
#ICLR2024
@gembioworkshop
paper submission deadline is coming up in ONE week: February 3rd! We have multiple tracks that should be of interest to both the ML community and experimentalists!
0
5
22
There’s really no better than feeling than when your first mentees get the
@NSF
GRFP!
@garykbrixi
, you’re going to continue to be a scientific rockstar in grad school! 🌟 You and
@SabrinaKoseki
are the most deserving recipients! I’m so proud of you guys. 💙
1
0
23
Residue energy-based preference optimization of antigen-specific antibodies. 🫨 A very interesting way to get around the lack of diverse paired structures for training!
0
1
23
Hi all, I'll be presenting our team's work on designing target-binding peptides with generative language models (and other cool protein design work!) tomorrow
@valence_ai
! 💻🧫 Please find Zoom details here: . Looking forward to seeing you there! 🥳
1
2
21
Beautiful beautiful work! I’ve said that pLDDT really tells you about how disordered proteins are — now we can leverage AF2 with simulations to explore it! 🥰 Would be excited to see this extend to AF2-Multimer as well. 😅
Happy to share the publication of our paper in
@Nature
Conformational ensembles of the human intrinsically disordered proteome
Work led by
@GiulioTesei
and
@AnnaIdaTrolle
. I'll post more later, but for now here is a link:
and a short movie about the work
53
315
1K
0
1
20
Really excited to try this out! Btw, we should make it a norm for protein language models to be hosted on HuggingFace! 🤗
🦅 the FAbCon has landed 🦅
Our
#generative
#antibody
#LLM
available on
🐣 FAbCon-small (144M)
🐥 FAbCon-medium (297M)
🦅 FAbCon-large (2.4B)
Team effort
@alchemabtx
@all_your_bayes
@aretasgasp
@jhrf
@jakegalson
@JorgeNDias
🎉
2
27
108
1
3
20
Very excited to have our tour-de-force genetic screening paper out in
@CellRepMethods
! We've developed a powerful, integrated pipeline for identifying, screening, and interrogating genetic perturbations for defined phenotypic outputs. Let's review how STAMPScreen works! [1/5]
When scientists at the Wyss,
@harvardmed
, &
@medialab
got frustrated w/ existing tools & methods for gene engineering, they made their own.
@PranamMIT
,
@krammecc
, & colleagues invented STAMPScreen to help scientists get from a database to results fast.
0
5
12
3
5
18
@andrewwhite01
Miniproteins are this weird intermediate between the specificity of an antibody and the permeability/drug-like properties of peptides. I personally think miniproteins are a thing because diffusion/flow-matching are not ideal for antibodies or peptides. At least with the de novo
0
1
17
@Pandeylab
@NatureComms
Do you understand how dangerous it is to draw strong conclusions from poorly analyzed data with severe methodological flaws? People look at journals such as
@NatureComms
for legitimate conclusions that can affect hiring, public practice, etc. This paper should be retracted!
1
0
16
An incredible paper that I have been excitedly anticipating is out in
@Nature
today -- so many congratulations to my brilliant friend and collaborator
@MartinPacesa
and the
@MartinJinek
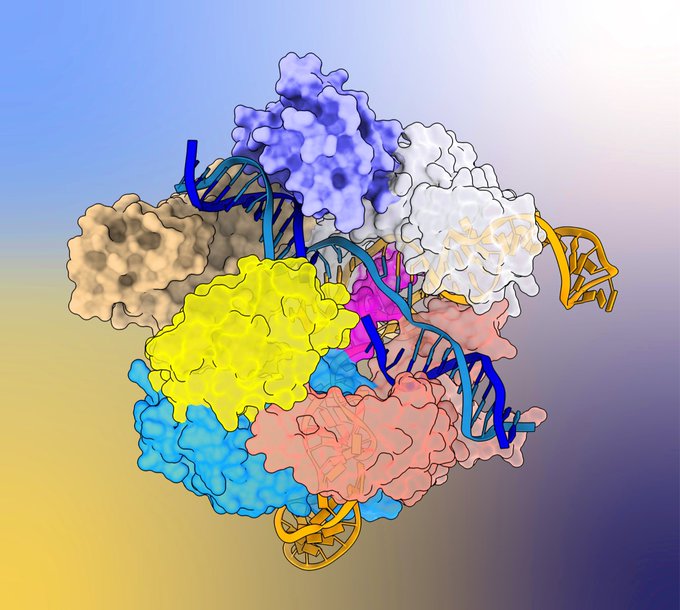
lab on this groundbreaking structural elucidation of Cas9 conformational activation! 🎉🎊🎈
Very excited to see our work on Cas9 conformational activation out in
@Nature
!
What started as a very poor crystal structure back in 2017 in the
@MartinJinek
lab, turned into a full blown cryoEM project during the pandemic.
30
73
530
1
0
16
Hi all! I hope you can take a few minute to learn about our latest research on designing target-binding peptides (amongst other useful proteins) using
#GenerativeAI
! Thanks so much to
@valence_ai
for having me! 🙌🏾
0
2
15
Check out our new COVID-19 diagnostic platform, out today in
@ScienceAdvances
! Peptide beacons + mini-TIRF technology = sensitive S-RBD protein detection at femtomolar resolution. Really a great extension of our ubiquibody design work! 🤩
2
2
14
Read more about what my company
@GametoGen
is up to: . It's crazy to see how far it's come since
@martinvars
and I founded it back in 2020! 🤯
@DinaRadenkovic
and
@krammecc
continue to lead it to newer heights -- you guys are amazing! 🌟
1
0
14
Super excited about trying out CellREADR from our friends in Josh Huang's lab here at
@DukeNeuro
! This will be very useful for dynamic cell state engineering applications. 🧫
0
0
13
Hi everyone! We are hosting Duke AI Day (sponsored by
@nvidia
) on our beautiful
@DukeEngineering
campus on June 7th!☀️If you work on developing or applying AI algorithms, we'd love for you to register (it's free to everyone!) and submit an abstract:
2
7
13
Okay, I know
#AutoGPT
is the AI hype right now, but check out the Regression Transformer! Discontinuous-masking-based autoregression for conditional protein sequence generation?! Yeah, my lab's definitely using this! 😎
1
2
12
Check out our early-stage work on designing computationally-optimized peptides that bind to SARS-CoV-2 and recruit E3 Ubiquitin Ligases for degradation! Could be a potential alternative to PAC-MAN?
#COVID19
2
2
12
With
#COVID19
raging through the world this winter, alongside promising vaccines from
@moderna_tx
,
@pfizer
, and
@AstraZeneca
, we present a novel antiviral platform for targeted intracellular degradation of SARS-CoV-2 via computationally-engineered peptide fusions. (1/4)
1
1
11
Absolutely beautiful work from my talented and amazing friend,
@JulesGrunewald
!! If you're crazy about base editing, what's better than doing both C->T and A->G at the same time?! 🧬🍾
2
1
11
@alexijielu
@francescazfl
@KevinKaichuang
@avapamini
@yisongyue
Many congrats to
@francescazfl
and the team on this beautiful work! 🌟 I will say that we've found in multiple cases that fine-tuning pLMs (specifically ESM-2-650M for de novo binder design and binding prediction work super well (and we've tested this experimentally).
2
0
11
Super exciting to see our initial idea of chimeric RNA:DNA guides () working well in the clinic! :) Excellent progress by
@CaribouBio
!
@njakimo
Very excited about these phase 1 results from
@CaribouBio
! The ultra high specificity of the chRDNA platform was crucial for achieving this. Very glad we had the opportunity to collaborate and elucidate the mechanism of increased specificity.
0
2
34
0
1
11
This is the incredible work of my brilliant, prodigious (brand new!) graduate student,
@pengzhangzhi1
. He just started in JANUARY and immediately innovated this beautiful architecture!
2
0
9
@Pandeylab
@NatureComms
If this was good science, then I'd be like okay, so be it and let's work to improve whatever conclusions we found troubling. But no, these were poorly drawn conclusions with no methodological or empirical basis.
3
1
9
Talk about the power of simulations — amazing to get fundamental mechanisms elucidated with MD! 🤩
A study in
@NatureChemistry
describes the discovery of glycan “gates” that open to allow SARS-CoV-2 entry. Read the paper:
9
162
446
0
1
9
#Llama3
is insane. 15 trillion tokens, 405 billion parameters with compute budget of 3.8×10^25 FLOPs. 😵💫 With that scale, I love the relative architectural simplicity: autoregressive decoder + DPO, with RoPE and grouped query attention built in. So clean. 🧼 I feel like we're
2
3
10
So many congratulations to my wonderful friend and collaborator, Dr.
@JooyoungLee10
!! What an incredible PhD thesis and defense!! So excited to have you in Boston!! 🎊🧬🎊
Congratulations to Dr. Jooyoung Lee for her successful defense of her outstanding Ph.D. thesis 👩🏻🎓 on
#antiCRISPR
! We will miss you but we know that you won’t be far away as you go on to do great things at
#Vertex
!
@JooyoungLee10
#CRISPR
@UMassMedRTI
@UMassMedical
#PhD
🍾🎉🧬
11
5
100
1
0
8
Incredible collaboration led by
@mpsmela1
and
@krammecc
! :) Very eager to see
@krammecc
and
@DinaRadenkovic
take this forward at
@GametoGen
! For a nice TL;DR, check out this highlight:
0
1
8
LIMA from
@MetaAI
is super exciting -- it shows us that well-curated, pre-trained LLMs are strong generalizers! For us protein designers, this is great news: we have solid pre-trained models (i.e. ESM-2) but limited, high-quality task-specific data. 🙌🏾
0
0
8
@hl_0826
I don’t see that happening. They basically are restricting anything with pharmaceutical value.
1
2
8
It's always a good day with FAIR and team come out with a new ESM model! The ability to generate novel sequences into desired structure holds huge promise for biologics and enzyme design -- excited for the team (
@r_manvitha
, et al.) to get started with it!
#proteindesign
Here’s what we learned from inverse folding on millions of
#AlphaFold
structures. Exciting time to bring a 800x new scale to
#proteindesign
. ESM-IF1 more accurately designs sequences to fold into desired structure, also unlocking new design capabilities.
9
78
326
0
0
7
Mixture-of-Depths! Such a cool, simple concept: dynamically allocate compute only to the most significant top-k tokens. This makes FLOP expenditure predictable, and can also serve as a great regularizer! Excited for the lab to try! Fun stuff,
@GoogleDeepMind
!
Paper:
0
0
8
This is the result of brilliant algorithm design by amazing undergrads
@bhat_suhaas
and
@kalyanmpalepu
, steadfast experimental validation by my PhD students,
@vivi_64_
and
@lauren_hong11
, and extensive computational benchmarking by the rest of the team. I'm a super lucky PI! ☺️
1
0
8
@sokrypton
@Patrick18287926
I love
@Patrick18287926
's work and we both work on peptide design (go peptides!), but I think this paper fell a little victim to the chicken and egg problem. It's really no one's fault, because they did the best with what they had in the PDB (and it's still a great study!):
2
1
8
Hi everyone! Just a reminder to register for Duke AI Day, which will be at
@DukeEngineering
on June 7th! 🤖 Please also feel free to submit a short abstract for posters/talks -- lots of great people, food, and science! More info here: See you there! ☀️
0
3
7
This paper was rejected, reviewed, rejected, reviewed again MULTIPLE times before finally finding a home at
@CommsBio
, one of my favorite journals! As a new PI, it was painful and rewarding, and I'm so grateful for my students and collaborators for staying the course together! 🥹
1
0
7
So much TRUTH. I'm thankful for such a positive and enriching PhD experience! I met so many wonderful people:
@njakimo
@JooyoungLee10
@ESontheimerLab
@BKleinstiver
@katieachristie
@JulesGrunewald
-- you guys made it worth it!! 🥰
0
0
7