Alex Rives
@alexrives
Followers
10K
Following
3K
Media
52
Statuses
331
AI for scientific discovery. Broad and MIT. Chief scientist at @EvoscaleAI and founder and scientific director of the ESM project previously @Meta.
Brooklyn, NY
Joined April 2009
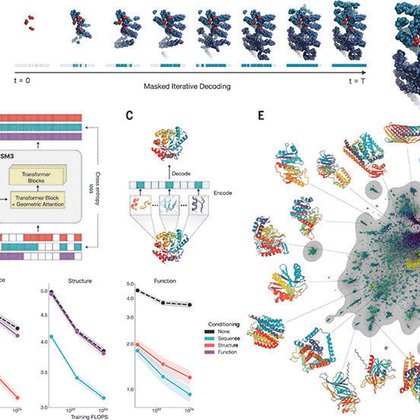
We're thrilled to present ESM3 in @ScienceMagazine. ESM3 is a generative language model that reasons over the three fundamental properties of proteins: sequence, structure, and function. Today we're making ESM3 available free to researchers worldwide via the public beta of an API
23
253
865
RT @pranamanam: Designing DNA-binding proteins shouldn’t require structures. ❌🧬 With DPAC, we align protein and DNA language models via con….
0
42
0
RT @davidrliu: In a medical milestone, a customized base editor was developed, characterized in human and mouse cells, tested in mice, stud….
0
433
0
RT @ScienceMagazine: Researchers have developed a deep learning protein language model, ESM3, that enables programmable protein design. Le….
0
165
0
RT @ScienceMagazine: An #AI model created to design proteins simulates 500 million years of protein evolution in developing a previously un….
0
245
0
RT @EricTopol: Whoa! When a large language of life model generates a protein equivalent to ~500 million years of evolution. @ScienceMagazin….
0
124
0
We’re also announcing an ESM3 compute grant pilot program for applications at the scientific frontier. ESM3 can enable creative applications of protein engineering from drug design, to green chemistry, and materials science. Please apply on Forge if your research could benefit.
We're thrilled to present ESM3 in @ScienceMagazine. ESM3 is a generative language model that reasons over the three fundamental properties of proteins: sequence, structure, and function. Today we're making ESM3 available free to researchers worldwide via the public beta of an API
4
21
102
RT @alexrives: Sign up for Forge, our new API for biological intelligence, now in public beta: .
0
11
0
RT @KevinKaichuang: We trained a model to co-generate protein sequence and structure by working in the ESMFold latent space, which encodes….
0
44
0
RT @amyxlu: 1/🧬 Excited to share PLAID, our new approach for co-generating sequence and all-atom protein structures by sampling from the la….
0
93
0
RT @afederation: This is what we've been waiting for. The best drug targets out there don't have structure, and won't anytime soon. The ke….
0
4
0
RT @eladgil: Evolutionary Scale · ESM Cambrian: Revealing the mysteries of proteins with unsupervised learning
0
8
0
RT @wolfejosh: Today @EvoscaleAI scaled up compute + data + trained next gen of protein language models. ESM C 300M + 600M w/open weights.E….
0
3
0