Alex Tong
@AlexanderTong7
Followers
3K
Following
350
Media
11
Statuses
189
Postdoc at Mila studying cell dynamics with Yoshua Bengio. I work on generative modeling and apply this to cells and proteins.
Montreal QC, Canada
Joined May 2017
RT @danyalrehman17: Wrapping up #ICML2025 on a high note — thrilled (and pleasantly surprised!) to win the Best Paper Award at @genbio_work….
0
11
0
RT @martoskreto: we’re not kfc but come watch us cook with our feynman-kac correctors, 4:30 pm today (july 16) at @icmlconf poster session….
0
15
0
Come check out SBG happening now! W-115 11-1:30 with.@charliebtan .@bose_joey .Chen Lin.@leonklein26 .@mmbronstein
0
18
91
RT @jacobbamberger: 🚨 ICML 2025 Paper 🚨. "On Measuring Long-Range Interactions in Graph Neural Networks". We formalize the long-range probl….
0
16
0
RT @bose_joey: 🚨 Our workshop on Frontiers of Probabilistic Inference: Learning meets Sampling got accepted to #NeurIPS2025!!. After the in….
0
3
0
Thrilled to be co-organizing FPI at #NeurIPS2025! I'm particularly excited about our new 'Call for Open Problems'track. If you have a tough, cross-disciplinary challenge, we want you to share it and inspire new collaborations. A unique opportunity! Learn more below.
1/ Where do Probabilistic Models, Sampling, Deep Learning, and Natural Sciences meet? 🤔 The workshop we’re organizing at #NeurIPS2025!. 📢 FPI@NeurIPS 2025: Frontiers in Probabilistic Inference – Learning meets Sampling. Learn more and submit →
0
2
25
RT @bose_joey: 🎉Personal update: I'm thrilled to announce that I'm joining Imperial College London @imperialcollege as an Assistant Profess….
0
34
0
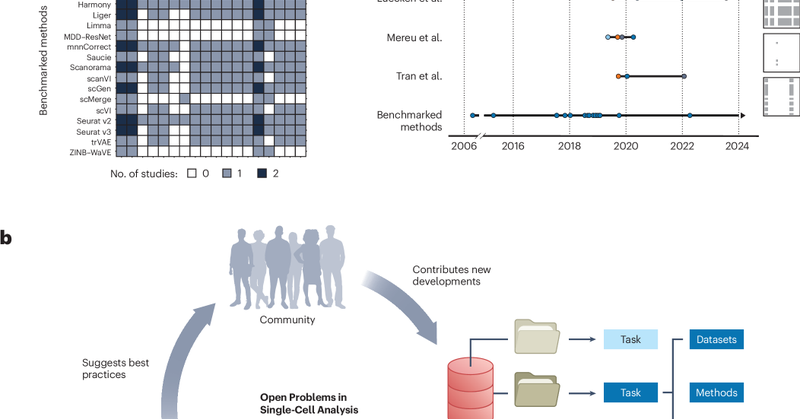
RT @fabian_theis: New OpenProblems paper out! 📝. Led by Malte Lücken with Smita Krishnaswamy, we present – a commun….
nature.com
Nature Biotechnology - Defining and benchmarking open problems in single-cell analysis
0
42
0
Yes! We were heavily influenced by your work @JamesTThorn although I think still quite challenging to train energy based models.
Nice to see energy based diffusion models + SMC/ Feynman Kac for temp annealing is useful for non-toy examples!.
0
0
7
RT @bose_joey: 🚨 I heard people saying that Diffusion Samplers are actually not more efficient than MD? . Well, if that's you, check out ou….
0
5
0
iDEM introduced a very effective but biased training scheme for diffusion-based samplers. This was great, but we were never able to scale it to real molecules.
arxiv.org
Efficiently generating statistically independent samples from an unnormalized probability distribution, such as equilibrium samples of many-body systems, is a foundational problem in science. In...
0
2
8
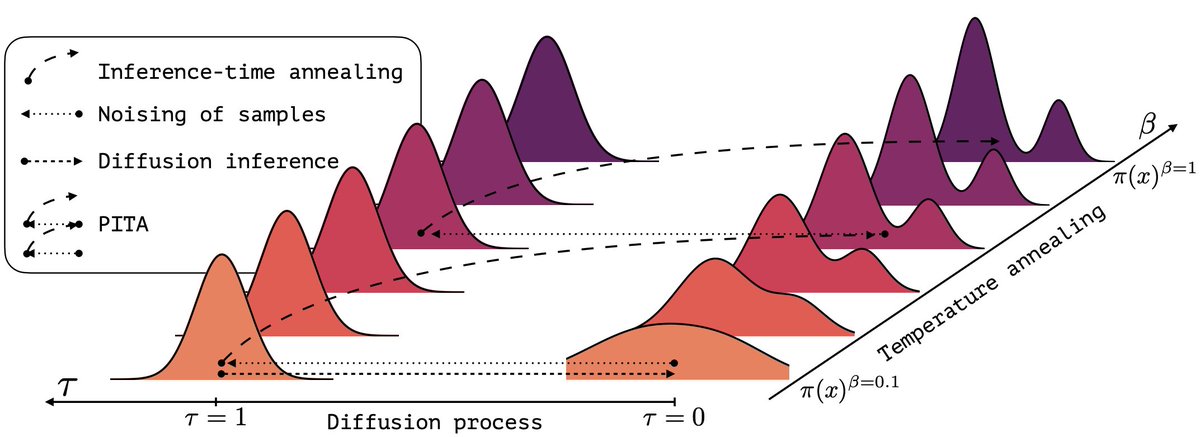
A bit of backstory on PITA: the project started with a key goal—to fix the inherent bias in prior diffusion samplers (like iDEM!). PITA leverages importance sampling to guarantee correctness. This commitment to unbiasedness is what gives PITA its power. See thread for details👇.
(1/n) Sampling from the Boltzmann density better than Molecular Dynamics (MD)? It is possible with PITA 🫓 Progressive Inference Time Annealing! A spotlight @genbio_workshop of @icmlconf 2025!. PITA learns from "hot," easy-to-explore molecular states 🔥 and then cleverly "cools"
2
4
34
RT @brekelmaniac: Given q_t, r_t as diffusion model(s), an SDE w/drift β ∇ log q_t + α ∇ log r_t doesn’t sample the sequence of geometric a….
0
6
0
RT @k_neklyudov: Why do we keep sampling from the same distribution the model was trained on?. We rethink this old paradigm by introducing….
0
26
0
Check out FKCs! A principled flexible approach for diffusion sampling. I was surprised how well it scaled to high dimensions given its reliance on importance reweighting. Thanks to great collaborators @Mila_Quebec @VectorInst @imperialcollege and @GoogleDeepMind. Thread👇🧵.
🧵(1/6) Delighted to share our @icmlconf 2025 spotlight paper: the Feynman-Kac Correctors (FKCs) in Diffusion. Picture this: it’s inference time and we want to generate new samples from our diffusion model. But we don’t want to just copy the training data – we may want to sample
2
5
34
RT @k_neklyudov: The supervision signal in AI4Science is so crisp that we can solve very complicated problems almost without any data or RL….
0
22
0
RT @bose_joey: Really excited about this new paper. As someone who spent a ton of time training regular flows with MLE and got burned FORT….
0
4
0
I'm particularly excited about the new opportunities this opens up for new fast architectures that are trained with regression but also have fast and accurate likelihood computation as heavily used in e.g. our work on Boltzmann Generators
arxiv.org
Scalable sampling of molecular states in thermodynamic equilibrium is a long-standing challenge in statistical physics. Boltzmann generators tackle this problem by pairing normalizing flows with...
0
2
15