Paras Verma, Ph. D
@parasvcb
Followers
212
Following
2K
Media
5
Statuses
655
Computational Biologist, Research fellow at IISER Mohali
Joined May 2012
Glad you like it! It's the nano-protein-viewer that I built over the summer Really need to work on marketing lol
@stevenyuyy Whoa what plugin is this? So much prettier than protein viewer
5
18
164
UF’s Perez Lab Gators team wins international AI protein design competition https://t.co/5GTPa9ZZk0
@UFChemistry
news.clas.ufl.edu
The Perez Lab Gators aim to produce high-impact, reproducible science through physics- and AI-based tools to improve human health.
1
1
21
RUNX1C isoform identified as driver of chemoresistance in acute myeloid leukemia - ... a way to potentially disarm it. In findings newly published in Blood Cancer Discovery, a team led by JAX assistant professor Eric Wang reports on ... -
news-medical.net
One of the biggest challenges in cancer treatment is that certain cancers reappear after chemotherapy-and an aggressive type of blood cancer called acute myeloid leukemia (AML) is notorious for this.
0
2
3
Thank You @biopatrika for featuring my work! Grateful to @ChabaRachna!
#authorinterview Dr. Swati Singh @swatisingh2511 working with Prof Rachna Chaba @ChabaRachna at IISER Mohali @IiserMohali talks about her work on "Decoding the impact of genetic variations in DgoR on D-galactonate metabolism" @JBacteriology @ASMicrobiology
https://t.co/d64iaWSGcZ
0
2
15
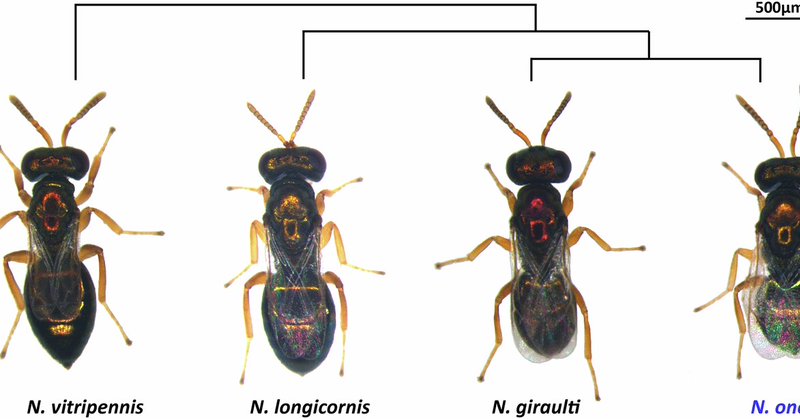
Happy to share our latest publication in Scientific Data @ScientificData on genome assembly and annotation of Nasonia oneida, the youngest species in the jewel wasp complex. @IiserMohali @DBS_IISERM
https://t.co/YwoWBWCafD
nature.com
Scientific Data - Genome assembly and annotation of the parasitoid jewel wasp Nasonia oneida
0
1
16
Postdoc position in our group at @emblebi , analyzing single cell data to better understand and treat neurodegen diseases, collab with @AndrewBassett43 , @mo_lotfollahi, @bayraktar_lab M Strauss and others @OpenTargets . Deadline 20/07. Please share🙏:
0
11
23
This is absolutely terrible. The benefits that those tools have brought to the scientific community are incommensurable.
2
18
61
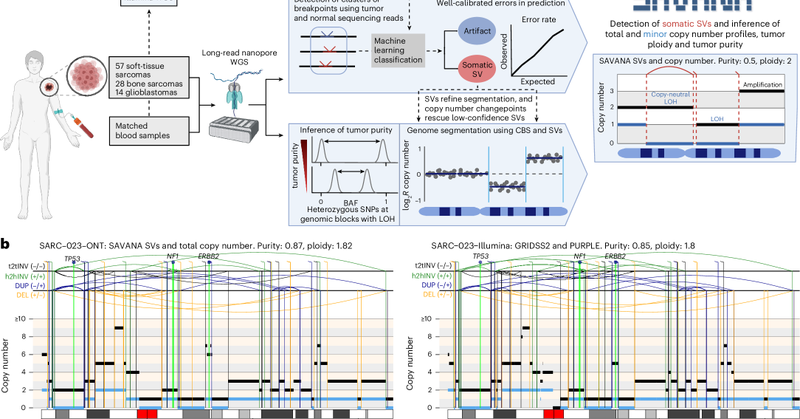
Thrilled to see #SAVANA out in @naturemethods🥳 SAVANA detects haplotype-resolved somatic SVs and copy number aberrations (SCNAs) and infers tumour purity & ploidy in clinical samples using long-read sequencing with or WITHOUT a matched germline control 👇 https://t.co/AdZhqHUsDb
nature.com
Nature Methods - SAVANA is a tool to detect somatic structural variants and copy number aberrations using long-read sequencing data, offering high sensitivity, specificity and compatibility with or...
3
28
134
Deep-learning-based single-domain and multidomain protein structure prediction with D-I-TASSER @NatureBiotech 1.D-I-TASSER is a hybrid deep-learning and physics-based pipeline that outperforms AlphaFold2 and AlphaFold3 in both single-domain and multidomain protein structure
2
34
136
"Only diamonds are made under pressure; good scientists are made with love and support." https://t.co/F77h4I6XYR
elifesciences.org
An eLife survey explores the experiences of those in the research community who support colleagues struggling with their mental health.
0
6
10
🧵 You filtered your VCF. But the output broke your pipeline. Why? You didn’t keep the header. Let's fix that.
2
5
54
New Online! Uncovering mRNA sequences that control translation initiation https://t.co/nfH6licvtD
0
2
7
A highlight of our recent research article by @_AndrewLeduc et al. https://t.co/3385ua1gGj
azolifesciences.com
A new study in Nature Communications presents a solution to protein leakage in single-cell proteomics, improving data quality by identifying and excluding permeabilized cells using a machine learning...
0
2
5
New online! RNA splicing — a central layer of gene regulation
nature.com
Nature Reviews Genetics - Technological and computational advances in recent years, from cryo-electron microscopy to sequencing technologies and machine learning, have substantially deepened our...
1
41
133
💪 What if we could map muscle repair pixel by pixel? 🔬 Our group leader Dr @Wi_Roman (@ARMI_Labs) built a low-cost spatial transcriptomics platform to do just that - revealing how muscles heal, grow & communicate. It's reshaping regenerative medicine. https://t.co/jZ3jMEosA6
emblaustralia.org
Dr William Roman, an EMBL Australia & Baker Foundation group leader at the Australian Regenerative Medicine Institute (ARMI), is reimagining the future of muscle repair – one pixel and protein at a...
0
4
12
14/14 📢 We welcome feedback, queries, and collaborations! Tag us or email to explore how ENACTdb can support your work. 📄 Method paper: https://t.co/1v48SjPflm 📄 DB paper: https://t.co/5rKVv1Nkv9
#AlternativeSplicing #Transcriptomics #Bioinformatics #Proteomics
academic.oup.com
AbstractMotivation. Gene transcripts are distinguished by the composition of their exons, and this different exon composition may contribute to advancing p
0
0
0
13🙏 Acknowledgments Thanks to my supervisor Dr. Shashi Pandit ( https://t.co/aO2jgJIJnB) for his constant mentorship. Also grateful to co-authors @Deeeksha_Thakur & @Deepanshi_awast for building the algorithm, DB, and analysis! Supported by @DBTIndia & @IiserMohali
#TeamScience
1
0
0
12/14 💻 Try ENACTdb now and find functional insights like: • Polymorphic attributes of exons • Mapping exonic variation to protein domains • Detecting how splicing reshapes protein features • Identifying indels/substitutions from exon changes. 🔗
1
0
0
11/14 🧩 Why use ENACT/ENACTdb? • Explore how splicing/transcriptional choices diversify the proteome • Understand how splicing architecture impacts local/global protein changes • Functional genomics, disease variant interpretation, evolution studies
1
0
0
10/14 🌐 ENACTdb: Data for the community and features! We annotated genes across 5 model organisms (human, mouse, zebrafish, fly, worm). ENACTdb enables: • Gene-based search (name or Entrez ID) • Exon-to-protein feature mapping (Pfam, structure, disorder) • Isoform comparison
1
0
0