Danny IncaRNAto
@incaRNAtolab
Followers
2K
Following
3K
Media
97
Statuses
2K
Associate Professor @univgroningen @ResearchGbb | Chief R&D Officer @serna_bio | #RNA structure ensemble dynamics | 🦋 https://t.co/VfgPl0Mwl0
Groningen, Netherlands
Joined December 2009
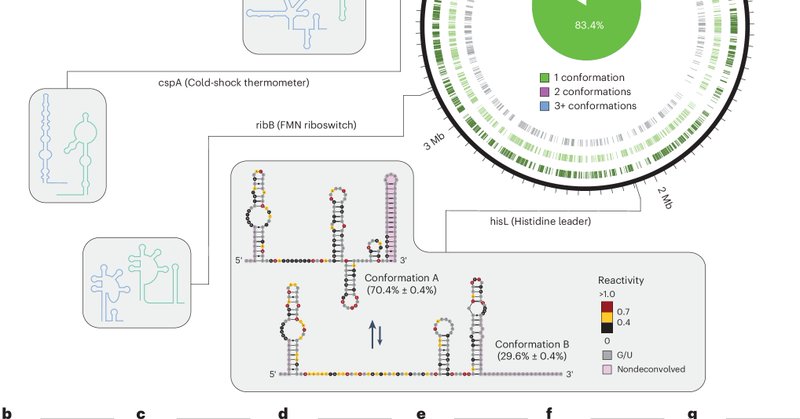
I am so incredibly excited to share our latest work, on the exploration of #RNA secondary structure ensembles and discovery of RNA regulatory structural switches in bacteria and human cells, out today in @NatureBiotech: https://t.co/tqiilEGJkp. A short tread! (1/n)
nature.com
Nature Biotechnology - Transcriptome-scale maps of RNA secondary structure ensembles in living cells detect candidate RNA structural switches.
8
44
187
As it's almost Christmas, here're 2 presents from the lab! 🎄 #RNA Framework 2.9.5 is out! Several additions, fixes and improvements. Check it out: https://t.co/XeMu0rj0o5 & see the CHANGELOG ( https://t.co/froquROCA7) for the details! The biggest improvement is the introduction
lnkd.in
This link will take you to a page that’s not on LinkedIn
0
2
10
We are pleased to host Prof Danny Incarnato on 5th December 9:30am @JohnInnesCentre. Please join us for this exciting talk. @YiliangDing @WHaerty @alperakay_ @EarlhamInst @uniofeastanglia @RNASociety @lexogen
0
4
9
New PhD opportunity available in my lab, in collaboration with Betty Chung (Cambridge), focusing on plant RNA viruses and AI-driven design.
findaphd.com
PhD Project - AI RNA Design for Developing Antiviral Solutions in Sugar Beet (Plant BioDesign JIC project) at University of York, listed on FindAPhD.com
0
11
22
!!! Important note: A bug was introduced in ViennaRNA v2.7.0, which breaks the pseudoknot detection functionality of rf-fold. However, RNAplot v2.7.0 is required to take advantage of the new secondary structure plotting functionality. Until a fix will be released, we advice users
0
0
0
Highlights: new normalization method in rf-norm (Mitchell method, https://t.co/iirIrWEKdm), statistics plots in rf-count & rf-count-genome, and secondary structure plots in rf-fold (powered by ViennaRNA v2.7.0) (2/n)
1
0
0
#RNA Framework 2.9.4 is out! Several additions, fixes, improvements. Check it out: https://t.co/7RKUitpX4p & see the CHANGELOG ( https://t.co/DZVj0o43ir) for the details! (1/n)
1
3
10
So excited to see this out in @NatureGenet! An amazing collaboration with the @gagneurlab I am happy I was (a small) part of... nucleotide dependencies can capture regulatory elements, including #RNA structures! Congrats to the whole team! Check it out:
nature.com
Nature Genetics - Mapping pairwise nucleotide dependencies by leveraging genomic language models highlights functional genomic elements and predicts deleterious genetic variants more effectively...
1
12
83
Looking forward!
Please join us for our next seminar, featuring Danny IncaRNAto, PhD: “Discovery of RNA regulatory structural switches via ensemble mapping” @incaRNAtolab #rna #casprnasig #rnastructure
0
1
5
CaCoFold-R3D: a probabilistic model that predicts RNA 3D motifs and secondary structure using evolutionary information. https://t.co/MLlxPadNDt
2
22
86
Happy to share that we identified a conserved 17kb long-range RNA-RNA interaction in SARS-CoV-2 that impacts virus fitness. Congratulations to all the authors! https://t.co/j4GBE6OU3W
nature.com
Nature Communications - Yang et al. apply high-throughput structure probing approaches to characterize the secondary structure throughout the SARS-CoV-2 WT and VOC genomes, uncovering an...
0
7
41
Termal: a fast and interactive terminal-based viewer for multiple sequence alignments. #MultipleSequenceAlignments #TerminalViewer #interactiveViewer #Bioinformatics @BioinfoAdv
https://t.co/WzGHLrKDYD
0
60
243
A great opportunity to work with one of the best. If you love #RNA and plants, this is for you!
0
2
11
Excited to have this work finally published on @NatureComms. With the Trcek lab @JohnsHopkins, @silvirouskin and @ThirumalaiArmy labs, we show that RNA structure impedes intermolecular base pairing in germ granule. https://t.co/1eAFcNLrxt
nature.com
Nature Communications - Tian et al. show that RNA folding limits extensive base pairing among mRNAs in Drosophila germ granules. Instead, mRNAs interact via scattered bases exposed on the surface...
0
4
35
I'm excited to share our new preprint on LagTag, a method that recovers both past and present chromatin states from the same mammalian cells. https://t.co/GEGQpN0Ff0
5
73
300
New online! Structural insights into higher-order natural RNA-only multimers https://t.co/KOjh7CNWSd
2
35
133
Excited to share our latest work with the bullock lab! We looked at how diverse mRNAs get selected for subcellular localization and it turns out that a single protein can recognize different RNA elements using shared features that weren’t apparent before. https://t.co/UG5is5elDm
1
13
59