Alberto Perez
@Al__Perez
Followers
1K
Following
2K
Media
21
Statuses
892
Asst. professor at UF. I use supercomputers to study DNA and proteins.
Joined November 2011
🚨 We’re Hiring! 🚨 @ASU_SMS Singharoy Lab is looking for an Applied Deep Learning Research Engineer to develop next-gen AI models for protein functional prediction. If you love building new deep learning architectures, pushing boundaries in model design, and collaborating with
0
3
9
We’re pleased to share our latest work focused on advancing FAIR data sharing for MD. A step toward making large-scale simulation data more accessible, reusable and interoperable for the community. #FAIRdata #MDsimulations
pubs.acs.org
The size of molecular dynamics (MD) trajectories remains a major obstacle for data sharing, long-term storage, and ensemble analysis at scale. Existing solutions often rely on frame subsampling or...
0
27
157
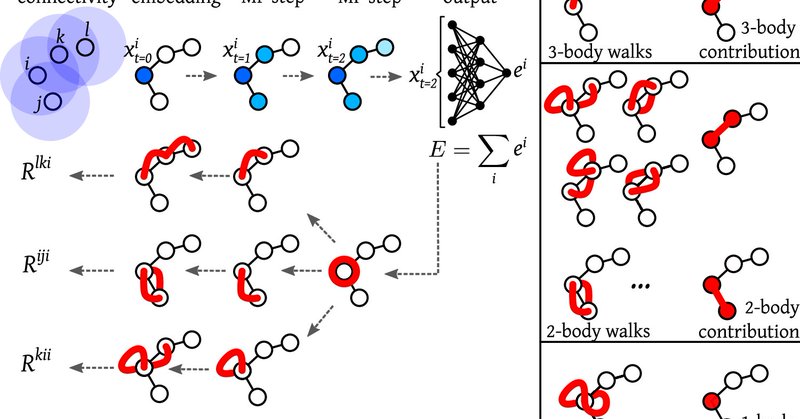
Interpretable #MachineLearning reveals the key physical interactions learned by coarse-grained MD models. By @CecClementi and team, led by Klara Bonneau. https://t.co/FSP3z4kUlY
nature.com
Nature Communications - Machine-learned force fields are becoming increasingly popular but suffer from their “black-box” nature. Here the authors adapt explainable AI techniques to...
2
31
169
(1/n) Can diffusion models simulate molecular dynamics instead of generating independent samples? In our NeurIPS2025 paper, we train energy-based diffusion models that can do both: - Generate independent samples - Learn the underlying potential 𝑼 🧵👇 https://t.co/TSurVY3YEl
12
141
839
After two years of work, we’ve made an AI Scientist that runs for days and makes genuine discoveries. Working with external collaborators, we report seven externally validated discoveries across multiple fields. It is available right now for anyone to use. 1/5
193
560
4K
MDZip: Neural Compression of Molecular Dynamics Trajectories for Scalable Storage and Ensemble Reconstruction 1. MDZip is a novel neural compression framework that achieves over 95% reduction in storage size for molecular dynamics (MD) trajectories while preserving essential
1
7
20
Great chance to share our vision for MDDBa towards training the next generation AI at the @UF AI Days symposium.
0
0
7
I think this is the most interesting/innovative part of BoltzGen. Diffusing to AF2-style encoding to co-generate both backbone and sidechains identities! 🤯
With this we train a model with the standard AF3 / Boltz-2 scalable architecture that has proven state-of-the-art for folding. Injecting conditioning inputs allows us to control the designed binder in various ways
1
33
237
Last call - open til Oct 30! Are you excited about #MachineLearning and developing new architectures for Molecular Biology? Joint us for the next chapter of BioEmu at @MSFTResearch AI for Science - Berlin DE or Cambridge UK. https://t.co/M4cNZrVo49
0
10
47
De-novo design of a random protein walker 🚀 New preprint from David Baker!🚀 1. A team of researchers has achieved a significant milestone in protein engineering by designing a random protein walker that diffuses along a designed protein track. This is a major step forward in
0
30
145
Today we’re launching Proteinbase, a single hub for experimental protein design data. Over 1,000 novel proteins are already live, each with computational predictions, experimental validation, and the method used to design them. Everything comes from one lab under standardized
7
51
191
Hiring for BioEmu project @MSFTresearch AI for Science. Berlin DE or Cambridge UK. https://t.co/M4cNZrVo49 - #MachineLearning Researcher https://t.co/19D9MpdeCY - Data Engineer https://t.co/72S24Pa5Yq - Applied Researcher https://t.co/cP3jwgOCoi - Principal Applied Researcher
BioEmu now published in @ScienceMagazine !! What is BioEmu? Check out this video: https://t.co/PAj96iKvR7
0
26
72
UF’s Perez Lab Gators team wins international AI protein design competition https://t.co/5GTPa9ZZk0
@UFChemistry
news.clas.ufl.edu
The Perez Lab Gators aim to produce high-impact, reproducible science through physics- and AI-based tools to improve human health.
1
1
21
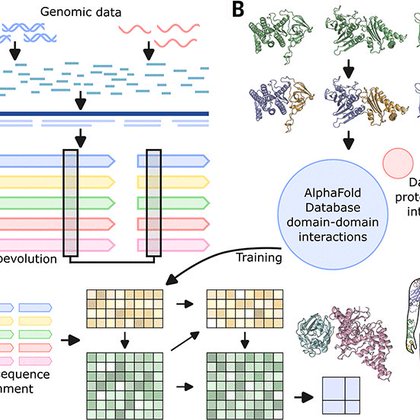
Thrilled to share that today we report our human protein interactome in @ScienceMagazine! Predicting which pairs of proteins in the human proteome is a great challenge due to the shallow eukaryotic evolutionary signals but hugely important to understand.
science.org
Protein-protein interactions (PPIs) are essential for biological function. Coevolutionary analysis and deep-learning (DL)–based protein structure prediction have enabled comprehensive PPI identific...
4
86
371
🧬La-Proteina🧬 The first generative model demonstrating accurate co-design of fully atomistic protein structures (sequence + side-chains + backbone) at scale, up to 800 residues, with state-of-the-art atomistic motif scaffolding performance - has just made its code open-source!
31
245
972
Very happy with our team at @UFChemistry for successfully participating in this Challenge. We learnt a lot and it was a great way to standardize pipelines and get everyone in the lab involved.
🚨The Bits to Binders Competition has concluded!🧬 One year ago we gathered scientists from around the world to design and submit protein binders that cause immune cells to target and eliminate CD20+ tumors Spoiler: They work!
1
2
21
🌟Women Make COMP @ ACS Spring 2026🌟 Celebrating the achievements of women & gender-diverse leaders in computational chemistry! 🌟 Open to all trainees 🌟 Promise in COMP Award 🌟 Free ACS registration + mentorship lunch w/ women leaders 👉Apply by Sept 29, 2025 @ link below
0
9
21
Exciting to see our protein binder design pipeline BindCraft published in its final form in @Nature ! This has been an amazing collaborative effort with Lennart, @csche11h, @sokrypton, @befcorreia and many other amazing lab members and collaborators. https://t.co/PTMoqQqwcU
19
167
721
I'm very proud to receive the @AmerChemSociety @ACSCOMP @OpenEyeSoftware Cadence Molecular Sciences Outstanding Junior Faculty Award in Computational Chemistry! Thank you also to @bobbypaton and @zoecournia. Congrats @JoshVermaas, Tibor and Denise 🥳 @chemistrytau @BioSoftTAU
12
5
92
✨ Headed to #ACSFall2025? Want some COMP stickers? Find an ACS COMP Executive Committee member and grab a few! 🧪💻🎉 #ACSCOMP #CompChem
0
1
18