JBacteriology Eds
@JBacteriology
Followers
3K
Following
638
Media
73
Statuses
565
Tweets from the Editors of the Journal of Bacteriology. Revived on August 4, 2022
Joined December 2015
The complex developmental mechanisms of nucleus-forming jumbo phages Mozumdar D, Agard DA, Bondy-Denomy J Curr Opin Microbiol. 88:102676, doi: 10.1016/j.mib.2025.102676 https://t.co/QSe2AA3lDa
https://t.co/t1mir9aoHs
pubmed.ncbi.nlm.nih.gov
Bacteriophages related to the jumbo phage ΦKZ (Family: Chimalliviridae) exhibit a complex developmental cycle. First, two large macromolecular compartments are assembled that surround and protect the...
0
0
0
George O'Toole @geiselbiofilm.bsky.social New in JB: Fung, Visick et al. show that a single SNP can impact biofilm formation and colonization of its squid host by V. fischeri.
journals.asm.org
Biofilms promote the attachment of bacteria to each other and to surfaces. For Vibrio fischeri, biofilm formation dependent on the symbiosis polysaccharide (syp) locus promotes colonization of its...
0
0
0
George O'Toole @geiselbiofilm.bsky.social New in JB: Antonia Herrero reviews the distinct aspects of cell division for Anabaena, a multicellular cyanobacterium https://t.co/r1H0NnMPxy
journals.asm.org
Cyanobacteria are oxygenic phototrophs that make an important contribution to the global primary productivity in the biosphere (1). Indeed, they are the organisms in which oxygenic photosynthesis,...
0
0
0
George O'Toole @geiselbiofilm.bsky.social New in JB: Pinho, Gotz & Peschel review the history of S. aureus as a model for studying bacterial cell biology and pathogenesis https://t.co/9UCDW8O3HU JB History of Microbial Model collection:
journals.asm.org
0
0
0
George O'Toole @geiselbiofilm.bsky.social New in JB: Rotaru, Kotoky et al. review the surface biology of Methanosarcina, an archaeon, with respect to their EET strategies, and biogeochemical and industrial roles.
journals.asm.org
Methanogenic archaea, including Methanosarcina, play crucial roles in biotechnology and climate processes, impacting wastewater treatment, carbon capture, and greenhouse gas emissions. Methanosarcina...
0
0
0
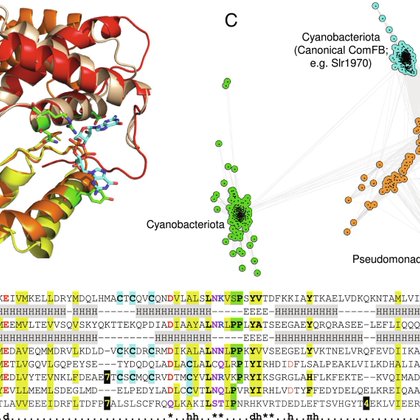
Over two years in the making, but here it is: ComFB, the third family of c-di-GMP receptor proteins with representatives in multiple bacterial phyla (PMID: 40966295) https://t.co/nC6hQx0WHR
pnas.org
Cyclic dimeric-GMP (c-di-GMP) is a ubiquitous bacterial second messenger that regulates a variety of cellular processes, including motility, biofil...
1
5
12
Rubisco is (arguably) the most abundant protein on Earth. (LPP surely comes close, right?) It’s an enzyme that fixes CO₂ into sugars during photosynthesis. Unfortunately, as most people learn in school, Rubisco is inefficient. Sometimes it confuses O₂ for CO₂ and wastes
39
394
2K
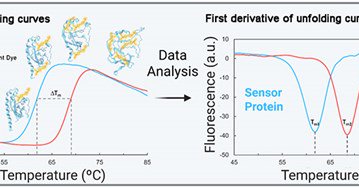
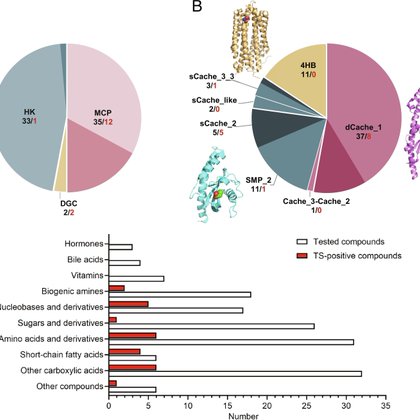
📷 @miguelmatilla.bsky.social How do bacteria sense their world? Our review summarizes thermal-shift assay approaches for identifying ligands for bacterial sensor proteins
academic.oup.com
The thermal shift assay has enabled major advances in the study of bacterial signal transduction and receptor function.
0
1
2
https://t.co/Yq7Bp2ppUy The PDZ domain of EpsC is required for extracellular secretion of VesB by the Type II secretion system in Vibrio cholerae
journals.asm.org
The T2SS is common in Gram-negative pathogens, facilitating the secretion of various toxins and enzymes; however, the mechanisms of substrate selection and secretion remain poorly understood. Here,...
0
2
2
Out Now! Pseudomonas aeruginosa senses exopolysaccharide trails using type IV pili and adhesins during biofilm formation https://t.co/72bvfUbLcl
#PseudomonasAeruginosa #BiofilmFormation #Microbiology
0
19
53
Lara Amorim, Conceição Santos and Kenneth Timmis Prioritising Microbiology in Secondary Education Addresses Emerging Scientific-Social-Educational Challenges and Competency Needs https://t.co/M7YhgHHnnQ
0
1
1
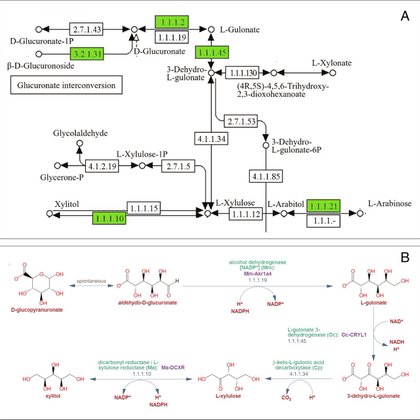
Some thoughts on metabolism and metabolic pathway holes https://t.co/1ZwExUprti.
pnas.org
Using bioinformatics for identifying and plugging metabolic pathway holes
0
3
10
In our next release (2025_04) we will deploy a new Reference Proteome selection pipeline with the aim of improving the representation of species biodiversity in UniProtKB. Read our blog post to find out how this might affect your research: https://t.co/7oGRdWFVbl
0
8
18
Which chemicals motile bacteria sense in the human gut? Great collaboration with @zhulinlab and @BangeBalcony, and congratulations to @wency13419273 and @EkaterinaJalomo! Specificities of chemosensory receptors in the human gut microbiota | PNAS
pnas.org
The human gut is rich in metabolites and harbors a complex microbial community, yet surprisingly little is known about the spectrum of chemical sig...
2
11
28
Our group review on the history of #Myxococcus xanthus as a model organism ( https://t.co/GdNIKQHAbf) made the cover of the Journal of Bacteriology!!!
0
3
19
Nuestro más reciente artículo "Deciphering the function of Com_YlbF domain-containing proteins in Staphylococcus aureus" publicado en @JBacteriology de @ASMicrobiology. https://t.co/Trb8q3MytW
1
1
2
Explore 🧭 a residue’s structural environment in the context of its linked annotations. Colour-coding 🖍️residues in the sequence points you to places of interest, go and discover. Try yourself: https://t.co/fQeBG5VcPW
#PDBeBeta #ResidueLevelInsights #PDBeValidation #ARISE #IUCr
0
7
14
JB editors' pick: Conjugative delivery of toxin genes ccdB and kil confers synergistic killing of bacterial recipients https://t.co/ToRxQJpwYp
journals.asm.org
The prevalence of antibiotic resistance emphasizes the need for alternative antimicrobial intervention strategies. We engineered Escherichia coli for conjugative transmission of plasmids encoding...
0
0
3
Soluble peptidoglycan fragments do not initiate spore germination but stimulate germination triggered by amino acids and nucleosides https://t.co/mqiuaF9gM0
journals.asm.org
Stimuli and mechanisms that underpin bacterial spore germination are fairly well characterized. The physiological route relies upon the interaction of various small nutrient molecules with receptor...
0
0
2