Francis O'Reilly
@FJ_OReilly
Followers
781
Following
5K
Media
34
Statuses
750
Structural systems biology at the NCI/NIH. Views are my own.
Frederick, MD
Joined July 2017
RT @Carter_Lab: Excited to share our latest work with the bullock lab! We looked at how diverse mRNAs get selected for subcellular localiza….
0
12
0
Time to get signed up for the next Symposium on Structural Proteomics! This time in beautiful Milan, 6th-8th October.
ssp2025.squarespace.com
0
0
5
RT @anangbhai: RIP Brian Wilson. Sharing BBC Music’s tribute to The Beach Boys “God Only Knows”
0
2K
0
RT @kellogg_liz: Despite the chaos and turmoil, our lab is blessed to have 3 new postdoctoral fellow openings and we would welcome scientis….
0
12
0
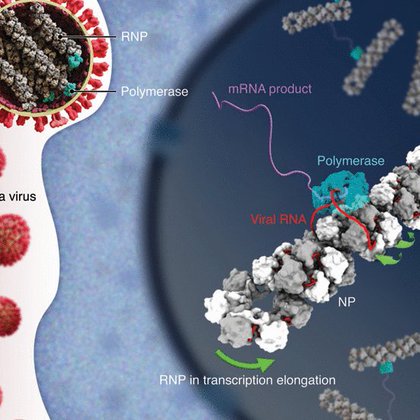
RT @yiweichang: Excited to share our new @ScienceMagazine paper! Led by postdocs Ruchao Peng and Xin Xu, we used cryo-EM/ET to reveal the i….
science.org
Influenza viruses replicate and transcribe their genome in the context of a conserved ribonucleoprotein (RNP) complex. By integrating cryo–electron microscopy single-particle analysis and cryo–elec...
0
26
0
RT @OlivierDuss: Ever wondered how several individual RBPs cooperate to repress translation?. To figure it out, we simultaneously tracked t….
0
9
0
RT @SergioCruzLeon1: Excited to share our pre-print on the molecular architecture of heterochromatin in human cells🧬🔬.w/ @jpkreysing, Johan….
0
120
0
RT @HaselbachLab: New Preprint: We have solved the structure of the proteasome in complex with two different ubiquitin chains: https://t.co….
biorxiv.org
Proteasomal degradation is a fundamental process for all eukaryotic life. A protein destined for degradation is first tagged with a polyubiquitin chain, which is selected by the proteasome. Different...
0
25
0
RT @HaselbachLab: we are looking for PhD students using time resolved cryoEM or cryoET on topics of the ubiquitin proteasome system. Howeve….
0
7
0
RT @fried_lab: Excited to share some work in our collaboration between @fried_lab and Ed O'Brien @psu_chemistry, just published in @Science….
science.org
Protein misfolding involving entanglements could be the structural origin of power-law distributions of folding times.
0
3
0
RT @BolandLab_GE: 🚨Preprint, RT appreciated🙏.Our work ‘Substrate recognition by human separase’ by @JunYu_unige, Sophia Schmidt, @Margheri….
0
24
0
RT @DanNomura: I’m a standing member on the Chemical Biology and Probes (CBP) NIH study section that was supposed to meet tomorrow. It just….
0
27
0
RT @StJudeResearch: Cuts in NIH research grants will have a lasting negative impact on science and medicine in the U.S. Grants will continu….
0
712
0
RT @deneke_v: Truly honored that our work was highlighted as one of many applications where #Alphafold has had an enormous impact. As some….
0
3
0
RT @ernstschmidd: Make publication ready figures from your #AlphaFold PAE files. Works on multimer and AlphaFold 3 files. Customize colors….
thecodingbiologist.com
Make high quality publication ready figures from predicted aligned error (PAE) files output by AlphaFold.
0
25
0
RT @naturemethods: .@theliulab @MRuwolt @diogobor @milanzord @absea_bio @Julia_RtaAn introduce a XL-MS standard based on human proteins or….
0
8
0
RT @fried_lab: Nevan Krogan @KroganLab @GladstoneInst @UCSF keynotes the first day of @SSP2024 with a manifesto on the universality of PPIs….
0
6
0
RT @fried_lab: #SSP2024 is almost here! Last thing to do is stapling together programs @broadinstitute with @FJ_OReilly and @Malvina_P….
0
3
0
RT @lapassmore: New work from the lab!.James Stowell led this project showing the importance of multivalency and phospho-regulation in mRNA….
0
43
0
RT @deneke_v: The life of each one of us began when a sperm and an egg came together. But what actually happens at a molecular level?.Our l….
0
101
0