Haselbach lab
@HaselbachLab
Followers
2K
Following
3K
Media
46
Statuses
943
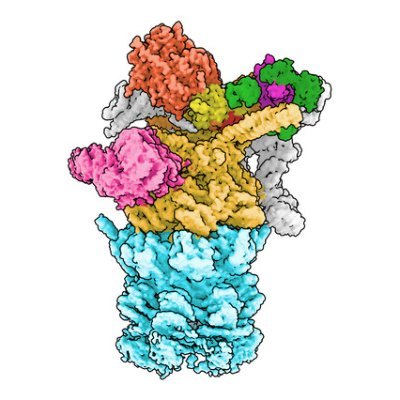
New Preprint: We have solved the structure of the proteasome in complex with two different ubiquitin chains: https://t.co/1TOgCKCYE1 This work spearheaded by PhD Student Sascha Amann Let me walk you through it 🧵 1/9
biorxiv.org
Proteasomal degradation is a fundamental process for all eukaryotic life. A protein destined for degradation is first tagged with a polyubiquitin chain, which is selected by the proteasome. Different...
1
24
92
How do cells decide on the life or death of a protein? 🔬 Researchers around @HaselbachLab at the IMP have captured the first high-resolution view of the human proteasome reading different ubiquitin signals. ➡️Read the full story: https://t.co/UIXBZogn5C
1
8
18
Similar results have been obtained earlier this year by @PDraczkowski et al. using a different kind of K11/K48 branched chain. See also their preprint: https://t.co/kNsg4NcRQo 9/9
biorxiv.org
Beyond the canonical K48-linked homotypic polyubiquitination for proteasome-targeted proteolysis, K11/K48-branched ubiquitin (Ub) chains are involved in fast-tracking protein turnover during cell...
0
0
4
This was a great team effort and we thank our Funders: @Boehringer, @FWF_at , WWTF, @ERC_Research , the @NIH. We also thank for the great support of the @IMPvienna and the help of the great facilities @VBCF_Tweets 8/9
1
1
6
Finally, we tested the interfaces genetically with our collaborators in the Zuber lab 7/9
1
0
3
The K11 branch uses a new binding interface between RPN2 and RPN10, further ubiquitins of this branch are pointing towards the solution and may not further interact with the proteasome 6/9
1
0
3
With our collaborator Nick Brown from @UNC we were able to generate K11/K48 branched ubiquitin chains that are the chains predominantly used for cell cycle control. 5/9
1
0
2
The 4th ubiquitin is thereby having the most interactions explaining why it is this one that boosts the degradation at least in vitro. 4/9
1
0
3
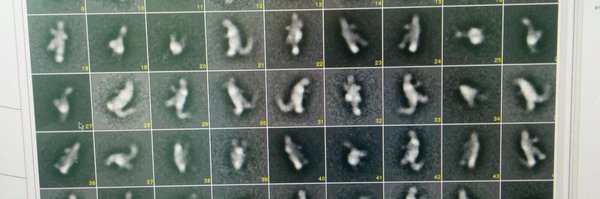
This enabled us to solve the structure of a ubiquitin chain bound proteasome with cryoEM. All four ubiquitins are clearly visible. Unexpectedly, we see the ubiquitin chain wrap around the UIM1 motif of the ubiquitin receptor RPN10 in a spiral like fashion. 3/9
1
0
3
we are looking for PhD students using time resolved cryoEM or cryoET on topics of the ubiquitin proteasome system. However, there is freedom to shape projects in own ways.
Passionate about structural biology and #biochemistry? Join David Haselbach’s group through the Vienna BioCenter #PhD Program (@TrainingVbc)! ⏳ Deadline: April 15, apply now: https://t.co/aB6BS6rREp 📷Watch David's 'Scientist Snapshot': https://t.co/4j99v9KQ4l
0
6
11
Passionate about structural biology and #biochemistry? Join David Haselbach’s group through the Vienna BioCenter #PhD Program (@TrainingVbc)! ⏳ Deadline: April 15, apply now: https://t.co/aB6BS6rREp 📷Watch David's 'Scientist Snapshot': https://t.co/4j99v9KQ4l
0
5
7
Congrats to @HannaBrunner_ PhD (@HaselbachLab), who recently defended her thesis: "The nuclear import of human proteasomes and the functional role of AKIRIN2". Well done Hanna! 🥂🍾 @univienna @IMPvienna @IMBA_Vienna @gmivienna @MaxPerutzLabs
0
2
13
Finally, my postdoc work is published in Cell Structure! 🎉 Grateful for the chance to apply deep learning to cryo-ET and learn from @HaselbachLab about structural biology🦠. Huge thanks to Lukas Herrmann for coding help and @GrohsPhilipp for my position and all the GPUs.
🔬Our @HaselbachLab with @HararPavol have developed ‘FakET’! The new method creates ‘fake’ electron microscopy images to train AI, reducing manual work in particle identification. Read the full story: https://t.co/ZeFW0WzwPu
#AI #ArtificialIntelligence #microscopy #cryoem
0
2
11
🔬Our @HaselbachLab with @HararPavol have developed ‘FakET’! The new method creates ‘fake’ electron microscopy images to train AI, reducing manual work in particle identification. Read the full story: https://t.co/ZeFW0WzwPu
#AI #ArtificialIntelligence #microscopy #cryoem
1
4
8
Extremely excited to be joining the vibrant community of the Vienna BioCenter to build up cryo-ET and cryo-FIB milling on campus soon!
IMBA and the @IMPvienna will soon host a joint research group headed by Sven Klumpe. The structural biologist will join both institutes with an innovative double-affiliation to add cryo-Electron Tomography research on “jumping genes” and other fields to the @viennabiocenter
1
1
34
🆕 #preprint by Karagoz lab & us on the crosstalk between #UFMylation & #RQC (ribosome-associated quality control) https://t.co/8RgM7yPxLJ As always let us know what you think!
0
11
31
Registration is now open for EMBO Workshop on Protein quality control: From molecular mechanisms to aging and disease 18 – 23 May 2025 in Hersonissos, Greece 📜 Early bird registration deadline: 31 Jan 2025 https://t.co/EelxCkiH1l
#EMBOproteinQualityControl
meetings.embo.org
Cellular protein quality control consists of a sophisticated network of molecular chaperones, enzymes and protein degradation machineries that tightly regulate all aspects of the life cycle of a prot…
0
3
4
This was only possible through amazing team work with our amazing collaborators the @PauliGroup from the @IMPvienna and @AndyMartinLab from @UCBerkeley We thank our funders @Boehringer and @WWTF for supporting our research (6/6)
0
0
3