Chengxiang Qiu
@CXchengxiangQIU
Followers
386
Following
154
Media
1
Statuses
20
Graduate student @uwgenome
Joined October 2017
Grateful to my mentors @JShendure, @coletrapnell, @CeciliaMoens, @thabangh, Bob Waterston, @KSusztak, and Qinghua Cui for their guidance and support along the way 🙏!!
0
0
5
Thrilled to share I’ve started my lab at Dartmouth’s Geisel School of Medicine! We focus on mapping cellular trajectories & TF networks in development and Mendelian disorders, exploring new therapies. Join us—postdocs, grads, and scientists welcome! https://t.co/8lHVDt3dHP
12
25
194
New paper from my lab and @JShendure lab! Led by the brilliant @Zukailiu and @CXchengxiangQIU. We tackle a question: How do anterior and posterior progenitor cells cooperate to self-organize into an embryonic structure (termed AP-gastruloid)? (1/n) https://t.co/PjZ7ZpuhOV
3
26
129
New synthetic biology / gene regulation lab opening in Zurich! We’re studying how to control cell state transitions – for example, making diseased cells healthy or creating new cellular functions – using systematic perturbations, single-cell genomics and machine learning. 🧵1/4
20
58
383
Incredibly proud to share that I recently joined @UCSF_BTS @UCSF as an Assistant Professor! This means that the Calderon Lab is now open! Or, more accurately, in the process of opening as I am still unboxing equipment and unraveling giant posters (a last @JSHendure lab gift) 🥳
19
27
193
Congrats,@CXchengxiangQIU, @bethkarenmartin, Ian, and @JShendure! What a fantastic paper which would be a long-years basis for the developmental biology field. "A single-cell time-lapse of mouse prenatal development from gastrula to birth".
nature.com
Nature - Single-cell transcriptome profiling of mouse embryos and newborn pups is combined with previously published data to construct a tree of cell-type relationships tracing development from...
0
9
44
Our latest out today in @Nature. We profiled 12 million single cells from mouse embryos spanning gastrulation to birth, defined cell type tree from zygote to birth, and unexpectedly found crazy fast changes within first hour of extrauterine life. OA PDF:
Excited to share our lab's latest preprint, led by @CXchengxiangQIU, @bethkarenmartin & Ian Welsh of @jacksonlab. We set out to build a single cell roadmap for all of mouse prenatal development, from single cell zygote to free-living pup. Preprint: https://t.co/GDS65Qf58X 1/n
15
153
581
Fantastic work delineating how life is generated by 12 million single-cell transcriptomes in a single study (from E8 to P0)! Congratulations to my former colleagues at @JShendure lab, especially @CXchengxiangQIU and @bethkarenmartin !
Excited to share our lab's latest preprint, led by @CXchengxiangQIU, @bethkarenmartin & Ian Welsh of @jacksonlab. We set out to build a single cell roadmap for all of mouse prenatal development, from single cell zygote to free-living pup. Preprint: https://t.co/GDS65Qf58X 1/n
0
7
31
Excited to share our lab's latest preprint, led by @CXchengxiangQIU, @bethkarenmartin & Ian Welsh of @jacksonlab. We set out to build a single cell roadmap for all of mouse prenatal development, from single cell zygote to free-living pup. Preprint: https://t.co/GDS65Qf58X 1/n
15
226
649
It is with great sadness that we share the news that Dr. Casey Brown passed away on Saturday, March 18. He was a friend, colleague, and mentor to so many in the GCB and CAMB communities, as well as the scientific community as a whole, and will be deeply missed.
43
37
181
It’s finally here! We built an artificial DNA-based recording system named DNA Ticker Tape – we can record the order of biological events in the genome, including those of the human cells! (1/12)
biorxiv.org
DNA is naturally well-suited to serve as a digital medium for in vivo molecular recording. However, DNA-based memory devices described to date are constrained in terms of the number of distinct...
Today we release in @biorxivpreprint a pair of related methods, DNA Ticker Tape & ENGRAM, for multiplex, temporally ordered genomic recording of cell lineage histories, enhancer & signal transduction activities, respectively led by the brilliant @_Choi_Junhong & @wchenomics 1/25
6
54
191
My interests in history makes me want to RECORD and WRITE cellular history too. How can we learn the past of a cell and understand why it is what it is right now? Can we record histories into genomes? Check out our latest preprint about ENGRAM https://t.co/Ogr2PEbroO
biorxiv.org
Measurements of gene expression and signal transduction activity are conventionally performed with methods that require either the destruction or live imaging of a biological sample within the...
Today we release in @biorxivpreprint a pair of related methods, DNA Ticker Tape & ENGRAM, for multiplex, temporally ordered genomic recording of cell lineage histories, enhancer & signal transduction activities, respectively led by the brilliant @_Choi_Junhong & @wchenomics 1/25
4
30
111
Our latest preprint, from the ingenious @bethkarenmartin, is an optimized, streamlined version of the three-level combinatorial indexing (split/pool) protocol for single cell RNA-seq (sci-RNA-seq3), originally developed by @junyue_cao. 1/4 https://t.co/HWFHNn6OB7
6
56
190
Our latest preprint, from the brilliant @CXchengxiangQIU, systematically integrates a series of single cell RNA-seq datasets that collectively span mouse gastrulation and organogenesis (E3.5 – E13.5) (1/4)
8
124
359
After >2000 commits, I am thrilled to announce the release of dynamo 1.0.0! In this release, we pushed the limits of our continuous vector field approaches to develop an awesome set of techniques to enable functional and predictive analyses of (time-resolved)scRNA-seq experiments
1
75
267
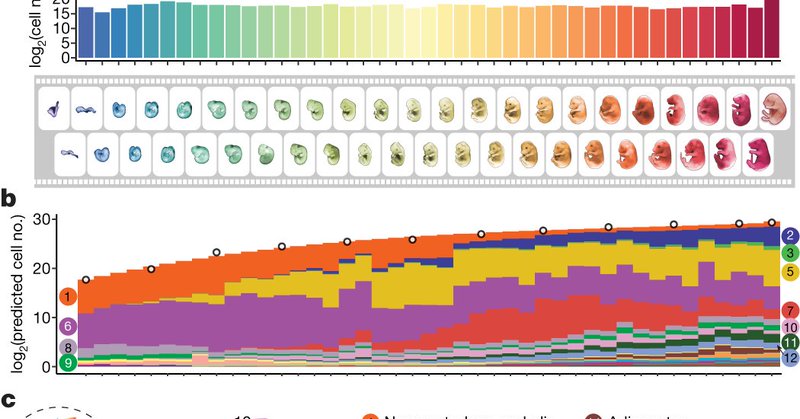
Excited to share a preprint about our project in which we aim to computationally reconstruct the cellular trajectories of mammalian embryogenesis, which started in my lab rotation at the beginning of my PhD. Many thanks to @JShendure and my rotation mentor @junyue_cao !!
Systematic reconstruction of the cellular trajectories of mammalian embryogenesis https://t.co/jLXmpGdo6Q
#bioRxiv
1
20
68
How do the ‘inner lives’ of cells in the early mammalian embryo change over time? Read this excellent N&V by @JShendure and @CXchengxiangQIU to find out https://t.co/mEWMYm4SNy
0
8
17
Very proud of Waddington's Waterslide, drawn by my 13 y/o daughter (for $5) as the original sketch for a @NatureNV with @CXchengxiangQIU on a terrific new paper from the Steltzer and Tanay groups at @WeizmannScience. https://t.co/S3QC8VuPfm
8
17
242
Our paper is out @NatureMedicine Renal compartment–specific genetic variation analyses identify new pathways in chronic kidney disease https://t.co/Yj2rQBXYMS Data is at https://t.co/vDbtff5KVO Congrats @CX_quorly
9
54
141