Ryan Tewhey
@r_tewhey
Followers
643
Following
694
Media
13
Statuses
395
Associate Professor at The Jackson Laboratory -- Studying gene regulation, genetic variation & human complex traits.
Bar Harbor, ME
Joined June 2016
RT @kanishkadey: Excited to share our latest preprint, as part of ENCODE4, on a consensus variant-to-function (cV2F) score for functionally….
0
48
0
RT @i_rodcast: Excited to share our sequence design work published in @Nature. I had the privilege of co-leading this effort alongside my f….
nature.com
Nature - A generalizable framework to prospectively engineer cis-regulatory elements from massively parallel reporter assay models can be used to write fit-for-purpose regulatory code.
0
23
0
RT @broadinstitute: Using #AI, a Broad/@jacksonlab /@YaleMed team has developed a way to decipher the language of genetic switches that tur….
0
35
0
RT @YaleGenetics: 🔥🔥🔥 new paper out from Steven Reilly @ReillyLikesIt together with Ryan Tewhey @r_tewhey led by Sager Gosai @SagerGosai +….
nature.com
Nature - A generalizable framework to prospectively engineer cis-regulatory elements from massively parallel reporter assay models can be used to write fit-for-purpose regulatory code.
0
8
0
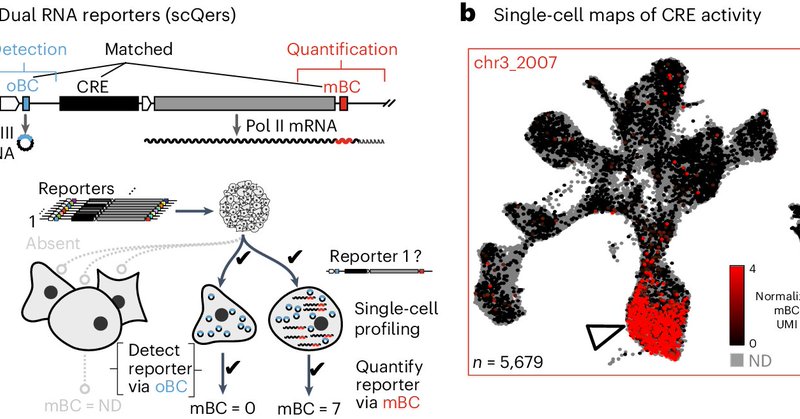
Fantastic scMPRA approach from @jb_lalanne, @SRegalad0 & @JShendure. Lots of great observations buried in the supplemental too!.
After a constructive set of reviews, our single-cell reporter work is OUT Check out the briefing if you want the tldr I am particularly fond of some of the new applications. Mini-update.
0
0
9
RT @jb_lalanne: After a constructive set of reviews, our single-cell reporter work is OUT Check out the briefing if….
nature.com
Nature Methods - We developed a two-pronged strategy to functionally probe the enormous repertoire of noncoding DNA within genomes. Our approach markedly improved signal-to-noise ratio and...
0
52
0
RT @jtung5: Two excellent sets of take-home impressions from @s_ramach and @ewanbirney on #BoG24. I'm going home excited and inspired by th….
0
11
0
Finally, we thank the @genome_gov for support. This work was funded by@ENCODE_NIH and data was released last year at @EncodeDCC. The project also allowed us to refine our MPRA protocol which we hope other groups will find useful for their own work.
0
0
3
@jacksonlab @ReillyLikesIt @LaylaSiraj @julirsch Data generation was the product of hard work and the expert hands of @KalesSusan, Danial (not on twitter) and @kousuke_mouri. We hope the data they produced will be valuable for many other projects going forward.
1
0
6
This was the first project we undertook when I started at @jacksonlab in Bar Harbor and a wonderful collaboration between the labs of @ReillyLikesIt and Hilary Finucane. Importantly, it was only made possible due to the careful work and brilliance of @LaylaSiraj and @julirsch.
1
1
9
I am delighted to see our work studying GWAS and eQTL causal alleles out in the wild. We learned several insights into how non-coding variation impart their effects. See Layla and Jacob’s tweetorial below for details.
Could not be more delighted to present our work investigating how over 220,000 complex and molecular trait-associated genetic variants affect transcriptional regulation using massively parallel reporter assays!.See below for a 🧵. 1/n.
1
2
21
RT @fuxmanlab: Had a great time hanging out with the @TewheyLab ! Thanks @r_tewhey and @jacksonlab for hosting us!
0
2
0
RT @anshulkundaje: This is by far the most convincing regulatory DNA synthetic design paper to date. Congrats to the authors 1/.
0
16
0
@jacksonlab @YaleMed @broadinstitute @i_rodcast @TewheyLab @SagerGosai @sabeti_lab @ReillyLikesIt Fun fact: The spark for this project came from experiments conducted by Natalia Fuentes during her summer stint in the @JAX_Education Summer Student Program. In 10 weeks she learned MPRA and produced amazing data that encouraged us to fully commit to this endeavor.
0
1
8
@jacksonlab @YaleMed @broadinstitute Shout out to @i_rodcast the brilliant computational scientist in the @TewheyLab who co-lead this project alongside @SagerGosai, and members of the @TewheyLab, @sabeti_lab & @ReillyLikesIt Lab for their help and support.
1
1
5
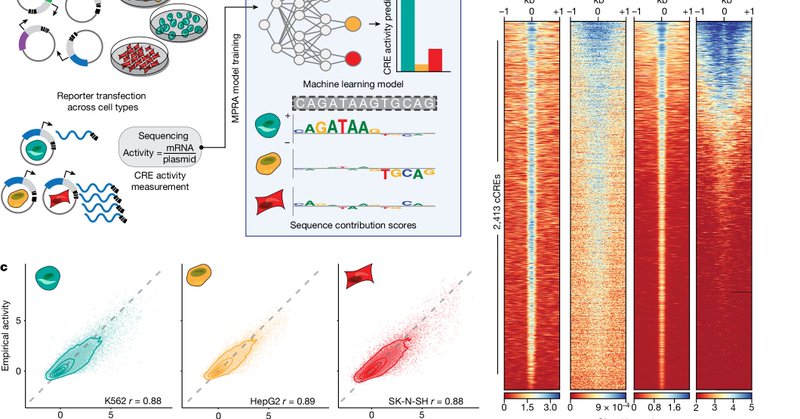
Five years ago, we wondered if we could design and validate synthetic, cell type-specific cis-regulatory elements that surpassed natural sequences. We are excited to share our successful approach, a result of a fantastic collaboration between @jacksonlab @YaleMed @broadinstitute.
I’m excited to share some work I co-lead with @i_rodcast and spanned the labs of @PardisSabeti, @ReillyLikesIt, and @r_tewhey to design and validate synthetic cis-regulatory elements with cell type-specific activity. 1/n.
3
15
73
RT @julirsch: Very excited to see our work on using gene features to identify target GWAS genes out at @NatureGenet.
0
5
0