Steve Reilly
@ReillyLikesIt
Followers
1K
Following
2K
Media
141
Statuses
936
Asst. Prof. @YaleGenetics making new experimental and comp tools to study gene regulatory functions of human variation. 🏳️🌈
Joined February 2010
Today @ ASHG 2 great posters from incoming postdocs to our lab Erin Gilbertson: 1030-Machine learning reveals the diversity of human 3D chromatin contacts Edwin Peña: 6054-Integrating ML and Functional Genomics to Prioritize CVD-associated Variants Altering Cardiac TF Function
2
2
7
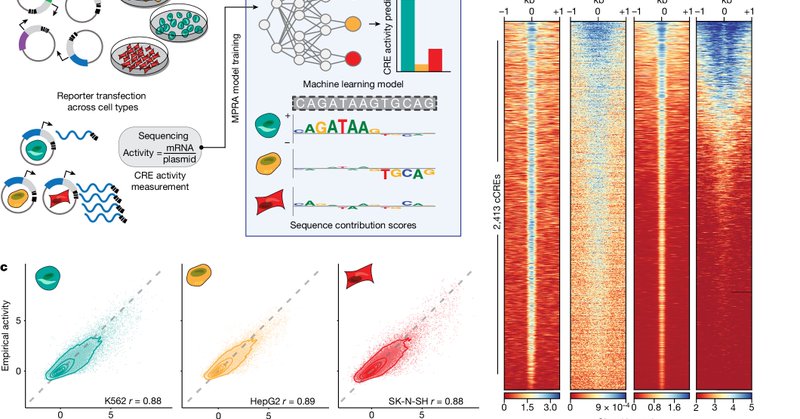
See John Butts's Poster #6047 Want to run an MPRA faster, cheaper, across cell types? We developed an accurate model of MPRA activity and highlight findings from assaying 100M+ variant effect predictions for fine-mapped variants, gnomAD, ClinVar, COSMIC, and more!
0
1
4
Here's an amazing explanation of our new paper from one of the brilliant minds behind it, @i_rodcast! Check it out.
Excited to share our sequence design work published in @Nature. I had the privilege of co-leading this effort alongside my friend and colleague @SagerGosai under the supervision of @r_tewhey, @ReillyLikesIt, and @PardisSabeti
https://t.co/mJSvyaMUHT 1/7
0
1
9
🔥🔥🔥 new paper out from Steven Reilly @ReillyLikesIt together with Ryan Tewhey @r_tewhey led by Sager Gosai @SagerGosai + Rodrigo Castro @i_rodcast Synthetic cis-regulatory elements (CREs) that drive cell-type-specific gene expression 🧬🤯
nature.com
Nature - A generalizable framework to prospectively engineer cis-regulatory elements from massively parallel reporter assay models can be used to write fit-for-purpose regulatory code.
0
8
58
Using #AI, a Broad/@jacksonlab /@YaleMed team has developed a way to decipher the language of genetic switches that turn genes on and off, and designed new, synthetic switches that control genes' expression in specific cell types. https://t.co/7XzesmAm92
@ReillyLikesIt
2
36
144
Awesome to be part of this effort, especially with @thanhthanhphd making a heroic push to go from somatic variants to function effects in just a few months!
Excited to share my lastPhD publication! We discovered non-inherited mutational patterns in schizophrenia occurring before birth. Huge collaborative effort w/ @seplyarskiy @jones_attila
https://t.co/yR9x5e8dLa
0
0
6
Our absolutely amazing postdoc @stephen_rong wrote a great review on MPRA, CRISPR, and machine-learning approaches to understanding the function of non-coding regulatory changes in human evolution. Take a look!
1
3
18
I am delighted to see our work studying GWAS and eQTL causal alleles out in the wild. We learned several insights into how non-coding variation impart their effects. See Layla and Jacob’s tweetorial below for details.
Could not be more delighted to present our work investigating how over 220,000 complex and molecular trait-associated genetic variants affect transcriptional regulation using massively parallel reporter assays! https://t.co/1eoN4OxAvd See below for a 🧵. 1/n
1
2
21
Click on replies in tweet 4 to see coauthor @julirsch continue the story!
1
0
3
Excited to release this 6+ year odyssey! I'm so proud to be part of the team of @LaylaSiraj @julirsch and the intrepid @r_tewhey who aimed our sights on one of the hardest problems in complex trait genomics. Read through this tweet-torial for the highlights and secret gems!
Could not be more delighted to present our work investigating how over 220,000 complex and molecular trait-associated genetic variants affect transcriptional regulation using massively parallel reporter assays! https://t.co/1eoN4OxAvd See below for a 🧵. 1/n
1
7
32
Exciting news from the department!
We are THRILLED to announce to appointment of our new Chair of the Department of Genetics @YaleMed, Dr Valerie Reinke. 👏🏽👏🏿👏👏🏼
0
3
12
Thanks to all the authors and labs who contributed to this work! Please reach out to us with any questions or thoughts with the paper. The data is all available now on the ENCODE portal.
1
0
0
From the very start of the human genome project, folks realized the genome was far too big for any one person to tackle alone. This is baked now baked into the field, with data sharing and collaboration a standard. It's my favorite way to do science.
1
0
0
This group made resources useful for the community: 1. screening guidelines ( https://t.co/VcNS6vMKl9) 2. identified best analysis methods ( https://t.co/rSVTbcrGO7) 3. share the best guides for targeting cCREs ( https://t.co/pXhnNIqW77) 4. 108 screens, 540K perturbations
1
0
1
This wouldn't have been possible without the 10+ labs labs in the @ENCODE_NIH consortium that provided data. These PIs and the folks @genome_gov, gave the resources, guidance/space to collaborate, and trusted a bunch of trainees to self-organize towards an important goal.
1
0
0
First off, to six (6!) co-first authors, were truly equals. They rejected the notion that this was 'splitting the pie' of credit, and decided to instead 'grow the pie' of the scientific impact. @davideyao
@JoshTycko
@JinWooOh6
@Lexi_Bounds
@SagerGosai
@LazLataniotis
1
1
1
I'll leave it to these amazing co-authors to explain the science. I'm so proud not just for this resource for non-coding CRISPR screens, but also demonstrating how productive collaborations can be when everyone is generous, creative, and motivated by the excitement of discovery.
Our new paper on non-coding CRISPRi screens to functionally characterize enhancers just came out in @NatureMethods ! https://t.co/UlKyTF3AvE
2
6
32
Our lab's Holiday Card tradition (adopted from the @sabeti_lab) continues into year 2. Can you spot the lab members and scientific jokes hidden in our growing laboratory? Check out https://t.co/FW6u0QDfwr for a key to all the science puns! Happy New Year from the Reilly Lab!
0
4
26