jingxin fu
@fu_jingxin
Followers
93
Following
149
Media
2
Statuses
54
Research Fellow @VanAllenLab - Ph.D. @XShirleyLiu lab. Genomics & Precision Cancer Medicine
Boston, MA
Joined March 2016
"I understand the gravity of being a woman and now a #Nobel laureate in the sciences. There aren't that many of us—yet." Prof. @CarolynBertozzi on chemistry, mentorship and representation ⬇️
56
1K
6K
Here's our latest - a deep molecular dive into one patient's journey to study the lasting effects of a severe environmental (chemotherapy) exposure over decades. Congrats @schianta_JL @Theodora_Pappa Dr. Park et al @JCOPO_ASCO @DanaFarber @DFCIPopSci
https://t.co/JKgRRmft09
ascopubs.org
3
25
87
My postdoc preprint in @XShirleyLiu lab! MetaTiME: Meta-components of Tumor Immune Microenvironment. Annotates cell type/states and signature continuum of TME scRNA, using an interpretable space learned from data and tailored for TME.
github.com
MetaTiME: Meta-components in Tumor immune MicroEnvironment - yi-zhang/MetaTiME
3
19
99
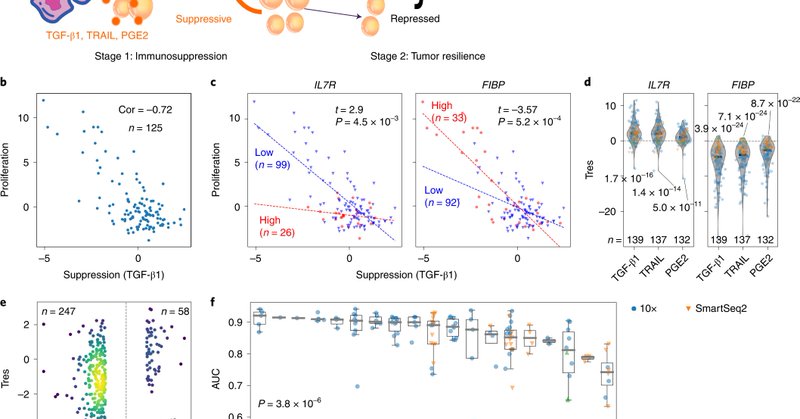
We are very excited to release our Tres framework to evaluate T-cell anti-tumor efficacy, and FIBP as a candidate immunotherapy target. The paper is published on Nature Medicine: https://t.co/mCLCIEwdcY The Tres server is available at:
nature.com
Nature Medicine - Gene signatures associated with T cell resilience to tumor-derived immunosuppressive signals predict responses to immune-checkpoint inhibitors and adoptive cell therapy and...
7
37
134
Interested in wet-lab single cell genomics and cancer? The @DanaFarber Center for Cancer Genomics (where I'm core faculty and longstanding collaborator) is hiring! Please share broadly: https://t.co/VVzUcUNfrD
0
32
62
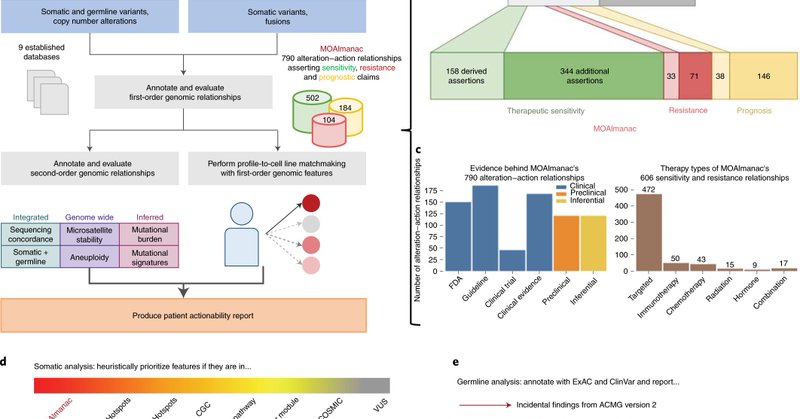
Our latest - introducing the Molecular Oncology Almanac (MOAlmanac): paired clinical interpretation algorithm + knowledge base for precision cancer medicine @moalmanac Congrats @brendan_reardon et al! @DanaFarber @broadinstitute
https://t.co/86JdStRgEi
@NatureCancer [1/12]
nature.com
Nature Cancer - Van Allen and colleagues develop a data-integration framework with an underlying knowledge base supporting clinical decision making and also serving as a hypothesis-generating...
5
68
218
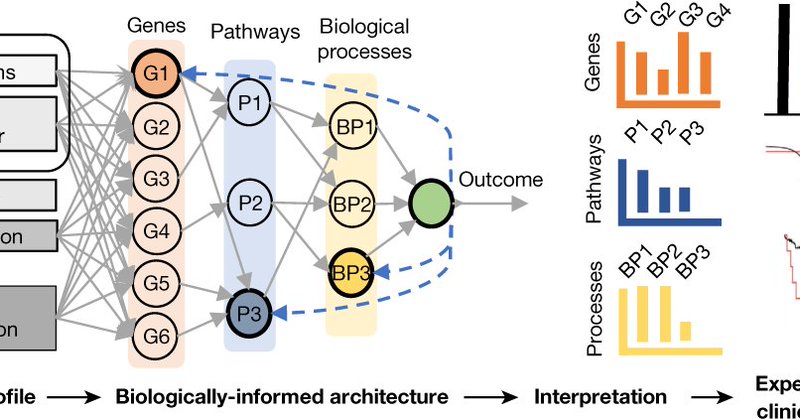
Here's our latest, where we explored the interface of biology and machine learning for cancer genomics discovery in prostate cancer - congrats @HMarakeby et al! @DanaFarber @broadinstitute
https://t.co/9Sba2htHfO [1/8]
nature.com
Nature - A biologically informed, interpretable deep learning model has been developed to evaluate molecular drivers of resistance to cancer treatment, predict clinical outcomes and guide...
14
161
490
Excited for our paper @NatureGenet on the landscape of molecular mechanisms regulating tumor cell response vs. resistance to natural killer cells #CancerResistance #NKcells #immunotherapy #CRISPR @DanaFarber @SU2C. https://t.co/NyUqXeYOSX. See thread below
7
30
110
Here’s our latest, a deep dive into fusions and partner or "collateral" cancer dependencies Interesting findings for pediatric and adult cancers... Congrats Dr. Riaz Gillani @BostonChildrens @DanaFarber @broadinstitute et al! @CR_AACR @AACR
https://t.co/iX6BlxIKWC [1/n]
3
25
87
congrats!
1/6 Our lab has developed the TRUST method to recover T/B-cell receptors from RNA-seq data. We present its new version TRUST4 in the @naturemethods paper, with features of better CDR3 accuracy, full-length assembly, and supporting single-cell data.
1
0
2
Immunotherapy resistance Single cell analysis Metastatic kidney cancer Here’s our latest - congrats Kevin @MXresearch @DrChoueiri et al! Lots to unpack: @Cancer_Cell @DanaFarber @broadinstitute @harvardmed
https://t.co/0y3nsGaE5T [1/n]
cell.com
Bi et al. dissect the cancer cell and immune microenvironmental transcriptional programs in immune checkpoint blockade-exposed metastatic renal cell carcinoma, revealing immune subpopulation reprog...
5
63
167
We are recruiting postdocs! Come join our multidisciplinary team to study fascinating computational biology! Multiple positions available. Please RT
0
10
17
Congrats @MXresearch !
Here’s our latest, a single cell dissection of lethal prostate cancer across metastatic sites & clinical states. Lots of cool stuff to unpack - congrats @MXresearch, Dr. Taplin et al! https://t.co/6K0gM9W4ZJ
@NatureMedicine @DanaFarber @broadinstitute [1/n]
0
0
5
Our paper describing ArchR, a single cell analysis platform designed for scalable analysis of open chromatin data, is out in Nature Genetics. Congrats to Jeff and Ryan! https://t.co/818T6S3ff5
2
116
426
Congrats!
Can deep learning reveal why certain mutations cause cancer? Why do oncogenes sometimes look like tumor suppressor genes? Our recent @MolecularCell paper provides answers and finds that altered protein degradation is a large contributor to tumorigenesis🧵 https://t.co/giM3bjYpNV
1
0
2
Can deep learning reveal why certain mutations cause cancer? Why do oncogenes sometimes look like tumor suppressor genes? Our recent @MolecularCell paper provides answers and finds that altered protein degradation is a large contributor to tumorigenesis🧵 https://t.co/giM3bjYpNV
8
77
209
TRUST4 is a great tool to analyze BCR and TCR sequences using unselected RNA sequencing data! It works for both single-cell RNA-seq and bulk RNA-Seq.
Great work from my colleagues!! Glad TRUST4 ( https://t.co/cYHgsHPkDL) is helpful in this project. Cross-site concordance evaluation of tumor DNA and RNA sequencing platforms for the CIMAC-CIDC network
0
3
3
I am excited to share a review that I wrote about building #infrastructure and #workflows for #clinical #bioinformatics #pipelines for Advances in #MolecularPathology. https://t.co/HBmdJY01Mt
1
14
44