Collin Tokheim
@ctokheim
Followers
524
Following
1K
Media
89
Statuses
729
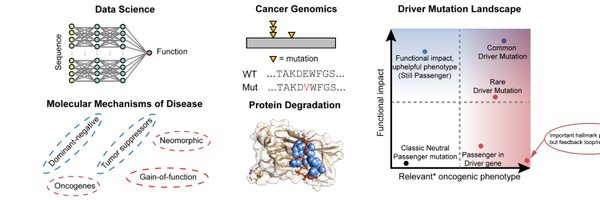
Topics: Cancer Genomics; Data Science; Targeted Protein Degradation. Prev: @DamonRunyon Fellow, @DanaFarber / Harvard; PhD @JHUBME. Opinions=own.
Boston, MA
Joined April 2018
Our paper on functional discovery of protein degradation and stabilization effectors with proteome-scale induced proximity screens is out in its final version! 1/ https://t.co/vzNbJyR2rk
nature.com
Nature - A synthetic proteome-scale strategy enables the identification of a diverse range of human proteins that can induce the degradation or stabilization of a target protein in a...
27
145
552
Exciting news! Our DFCI-TPD webinar channel reached 100,000 views on YouTube! Huge thanks to all speakers who shared their expertise, and all viewers! None of this would have been possible without our exceptional team @kdonovan1008 @RPNowak @BLZerf6 @hojong_yoon @MilkaKostic
2
15
66
In a packed room #ACMGMtg23 with @HeidiRehm Les Biesecker and Steve Harrison presenting the next version of variant classification work at @TheACMG @ClinGenResource AMP and CAP. Version 4 will be points based. All slides available at
2
37
99
Our lab website is online @MDAndersonNews ! https://t.co/eWM71CRcJh We're seeking highly motivated students, postdocs, and research assistants pursuing a career in cancer immunology and/or computational biology. Don't hesitate to apply!
mdanderson.org
The Gu lab is interested in enhancing the efficacy of cancer therapies, especially immunotherapy, by modulating cell-cell interactions.
0
23
55
ML marches toward TPD. Degradability correlates with ubiquitination potential & E2-accessible lysines in this model. It‘s fairly well appreciated now that formation of a ternary complex is necessary but not sufficient. https://t.co/YtHwV3dbjl
0
1
19
1/ My new book 'From Cell line to Command line' is finally here https://t.co/R4bsCE6Io8 Grab it to start transforming yourself to a computational biologist! A thread on how this book came into being: 👇🧵
30
187
981
So happy to see publishing of my doctoral work on targeted protein degradation. I’m very grateful to my advisor @XShirleyLiu for being the best mentor. Thank @ctokheim for all mentoring and help. Also thank @ssroyburman and @kdonovan1008 @eric_fischer lab and all coauthors!
https://t.co/Ra88NnTMEW This is our last piece of work on #proteindegradation at @dfcidatascience . Congratulations to @Wubing44589261 and @ctokheim , and thanks @eric_fischer Lab for the collaboration!
5
2
29
From the RF paper: "In an important paper on written character recognition, Amit and Geman [1997] define a large number of geometric features and search over a random selection of these for the best split at each node. This latter paper has been influential in my thinking."
0
0
0
While Leo Breiman rightly deserves credit for Random Forests in machine learning, the idea of randomized decision trees where a random subset of features are used actually came earlier by Amit & Geman:
1
1
2
Takeaway: Yes, Machine Learning is helpful for predicting pathogenic variants. But use the right method (VEST4, MutPred2, BayesDel or REVEL) with the right threshold!
0
1
5
Three methods (Sift, PolyPhen2, and CADD) had their own developer-recommended thresholds. "[these] tools classified a substantial fraction of variants in the gnomAD set as damaging (50.4% by SIFT, 29.3% by Polyphen-2, and 65.1% by CADD) ... suggesting a high false positive rate"
1
1
4
4 methods (VEST4, MutPred2, BayesDel, and Revel) could achieve the "strong" PP3 criteria for pathogenicity.
1
1
2
Pejaver et al. provide a very useful table on the score thresholds (intervals) for each method to support different levels of evidence. Note: PP3 and BP4 refer to guidelines by the American College of Medical Genetics and Genomics
1
4
2
"posterior probability" is just the probability that a variant is pathogenic (or benign) given the evidence (score) from each ML method on an independent dataset. Some methods at high scores provide "strong" evidence for pathogenicity, while others cannot be used at any score:
1
1
2
Since machine learning methods will typically output a single numerical score, Pejaver et al. calibrated each method's score to different clinical ACMG/AMP evidence levels that support classification as either a pathogenic or benign variant
1
1
3
How accurately can Machine Learning classify missense variants as pathogenic? A must read: https://t.co/YWpBbxXBi3 TLDR; There are good methods (BayesDel, MutPred2, REVEL, VEST4) that provide strong support, but the most commonly used (Sift, Polyphen2, CADD) have many FPs
1
24
78
@ctokheim @tycheleturner Efforts to make these datasets more accessible for variant curation are ongoing. We're working with @ClinGenResource to help make this happen, and @varianteffects is leading the way in defining standards and practices.
0
1
3
We are hiring! We are looking for an experienced scientist to join the Hematology Translational Medicine R&D group at @AstraZeneca in Waltham, MA as Senior Scientist, Heme Bioinformatics. Feel free to reach out with questions and apply here:
linkedin.com
Today’s top 8,000+ Senior Scientist jobs in United States. Leverage your professional network, and get hired. New Senior Scientist jobs added daily.
0
1
1
That being said, it does quite accurately hit a lot of the problems in the ML literature. And it is not just ML-specific venues, as I've seen a lot of these issues appear in ML methods published in the journal Nature.
0
0
2