Jakub Chrustowicz
@chrustowicz_j
Followers
122
Following
228
Media
3
Statuses
34
Postdoc at @MPI_Biochem | Schulman lab dissecting molecular mechanisms of GID/CTLH E3 ligases
München, Bayern
Joined June 2019
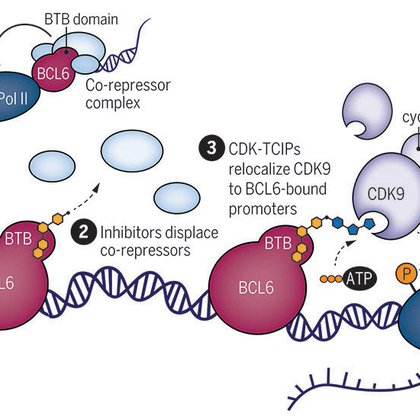
Thrilled to share our latest preprint describing discovery of the charged, metabolically activated molecular glue degrader engaging the basic pocket of the structural CRBN homolog, YPEL5 - a subunit of the giant GID/CTLH E3 ligase!!! https://t.co/XcAdbj7PNZ
2
18
65
Principles of paralog-specific degradation engaging the C-degron E3 KLHDC2, now out @SpringerNature @NatureComms Awesome collaboration with @Richard_E_Lee and Taosheng Chen labs. @StJudeStructBio @MPI_Biochem @StJudeResearch #TPD #PROTAC #SchulmanLab
https://t.co/FqdbKTGiaR
1
10
29
Can we turn the activity of oncogenic kinases against cancer itself? In my postdoc work @Stanford, new in @sciencemagazine, we show exactly that. https://t.co/PZjQxvab7z
#ScienceResearch #InducedProximity @Stanford_ChEMH @StanfordCancer @snsf_ch @StanfordMed
science.org
Kinases are critical regulators of cellular function that are commonly implicated in the mechanisms underlying disease. Most drugs that target kinases are molecules that inhibit their catalytic...
8
38
190
🔬 Neat discovery in protein degradation. Gray, Schulman, et al labs unveil ZZ1, a prodrug for charged molecular glues that target disease-causing proteins via the GID/CTLH ubiquitin ligase complex.
biorxiv.org
Small molecules that induce protein interactions hold tremendous potential as new medicines, as probes for molecular pathways, and as tools for agriculture. Explosive growth of targeted protein...
1
10
58
A great collaborative effort between @zhezhuang7, @WoongSubByun and Stephen M. Hinshaw at the Gray’s lab @Stanford, @StanfordMed, the Schulman lab @MPI_Biochem, and @zkozicka and Mikołaj Słabicki @biomiko at the Ebert’s lab @DanaFarber, @HarvardMed, @BroadInstitute!!!
0
0
5
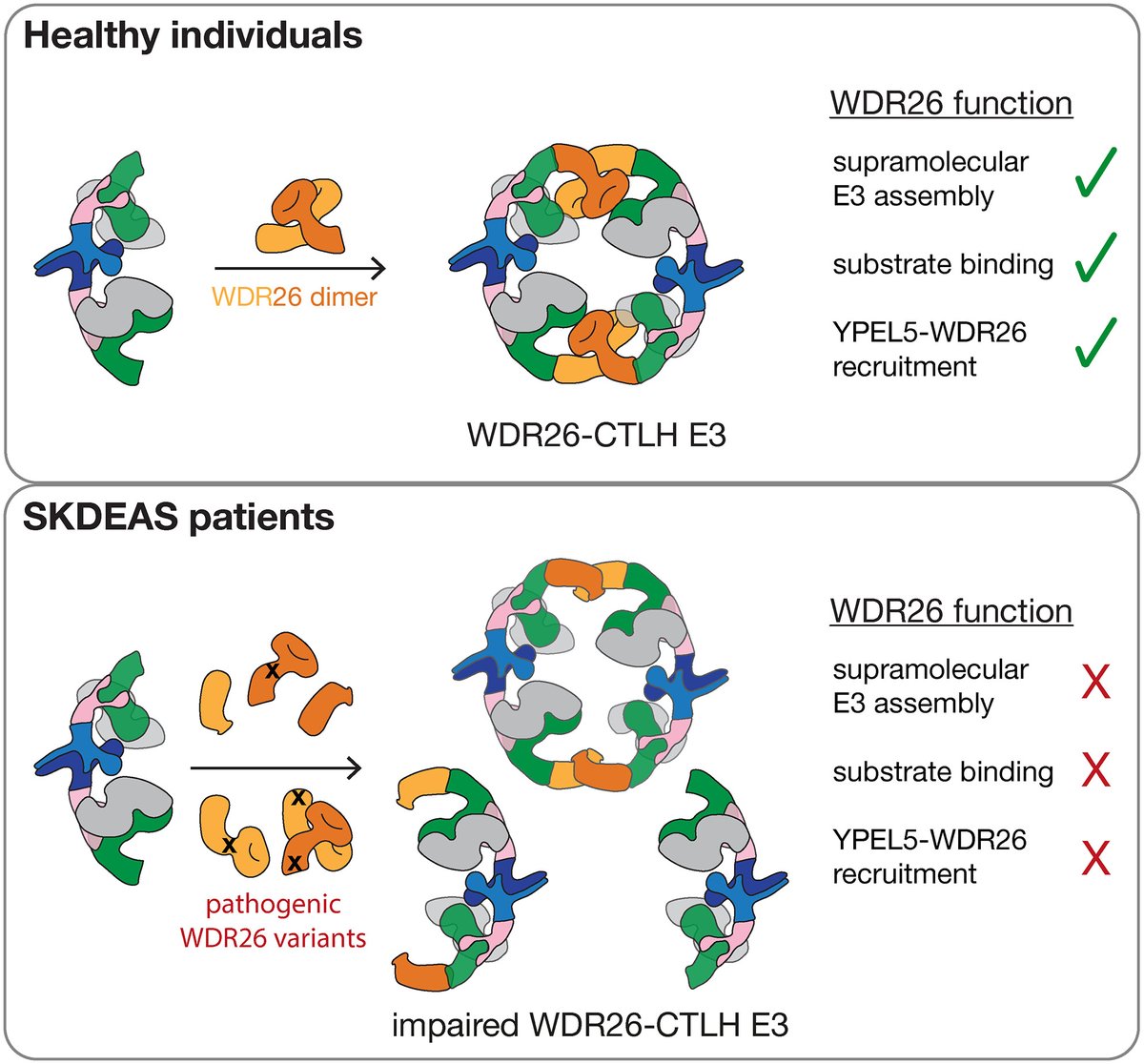
⭐️ Editor's Choice article: Mutations in WDR26 associated with #SkrabanDeardorffsyndrome (#SKDEAS) impair CTLH E3 complex assembly ➡️ https://t.co/oDwLlLWi6W
@pjmurray_lab @MPI_Biochem
1
3
7
Check out our story revealing exciting regulation of the NAD+ biosynthetic enzyme NMNAT1 by the supramolecular CTLH E3 ligase assembly out in @MolecularCell! ➡️ https://t.co/Fn3Hx7o38o Karthik Gottemukkala, @dawafutisherpa, @ArnoAlpi Schulman lab @MPI_Biochem
1
11
23
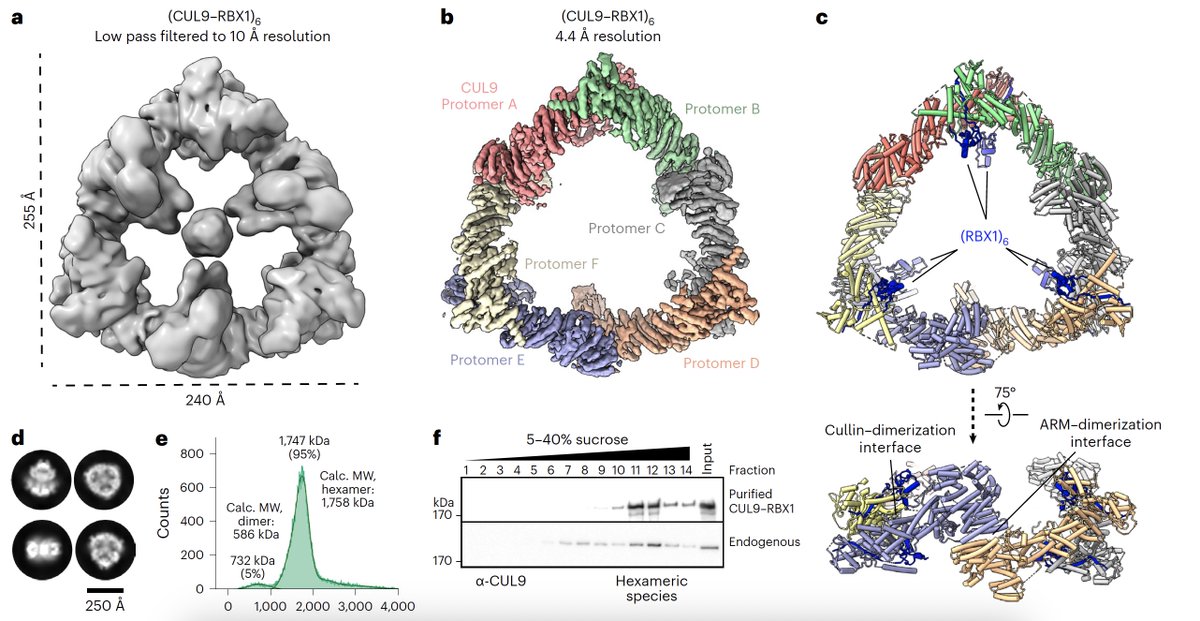
Wow! CUL9 is a GIANT E3 ubiquitin ligase! See unique cullin-RING-RBR all-in-one. +/- NEDD8. Now online in Nature Structural & Molecular Biology by Schulman Department ❕ https://t.co/G9y74fJONK
#ubiquitin #E3ligase #cryoEM
@DHornGhetko @LinusHopf @labs_mann @NatureSMB
0
21
76
🚨Excited to share that our manuscript is now published in @NatureSMB. Using #cryoEM 🔬we obtained multiple snapshots 📷 of the #chaperone-mediated multistep assembly pathway of the human 20S #proteasome. @MPI_Biochem @HarvardCellBio @ASAP_Research
https://t.co/1oJ1hiIRbR
2
13
33
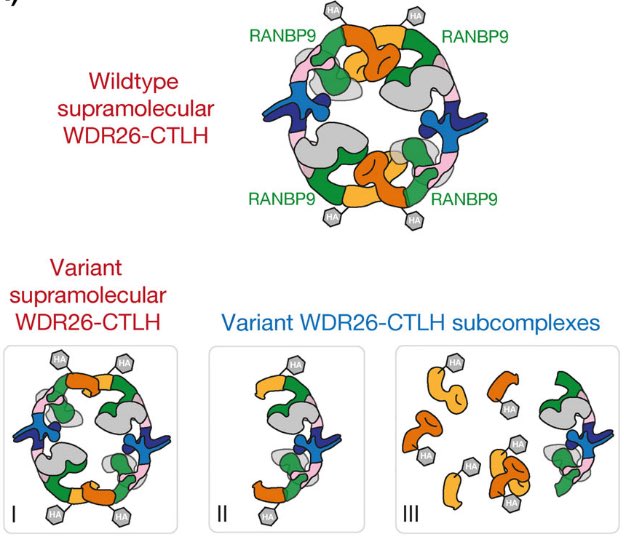
Here is the link to our SKDEAS paper: 10.1002/1873-3468.14866. And I love this figure. Maybe useful for the @Skdeas.org effort.
0
1
6
Now out in @MolecularCell! High-resolution structure showing direct ubiquitylation of a #TPD substrate! And much more, check out 👉 https://t.co/fZcP5DZcFf
#cryoEM @MPI_Biochem @UNLV_CoS @StJudeResearch @DanielS20127197 @rajanprabu (1/2)
4
15
80
How does multisite #phosphorylation configure the catalytic E2-E3 assembly for metabolic enzyme targeting? Check out our story available now in its final version in @MolecularCell! ❕ https://t.co/NjETPtjl7b
@chrustowicz_j @dawafutisherpa #ubiquitin
0
8
21
How does multisite #phosphorylation of an #E2 enzyme dictate its pairing with a multisubunit #E3 #ubiquitin ligase to regulate metabolism? Check out our story available now in @MolecularCell! ➡️ https://t.co/rAQe0E9Bju
@dawafutisherpa Schulman lab @MPI_Biochem
5
22
94
Which cullin-RING E3 #ubiquitin ligase directs your favorite pathway or your #DegraderDrug? Our conformation specific antibody and probing of active #CRL E3 complexes is in @nchembio! ❕ https://t.co/WST68AtFr1 ❕ https://t.co/uOo8UaA9uj
@labs_mann @pjmurray_lab @LukasHenneberg
0
7
28
In preparation for the Louis-Jeantet Prize ceremony, we accompanied the @LouisJeantetFDN video team to shoot parts of the video about our director Brenda Schulman, @iDikic2 and their amazing research in the #ubiquitin field. Watch the full video here: ➡️
0
3
8
Systemwide multiprotein complex formation visualized! CAND1 recycles Cullin-RING complexes to produce E3 #ubiquitin ligases. Averts cellular supply chain problems, prevents buildup of unneeded molecular machines & rapidly establishes degradation pathways ➡️ https://t.co/k7El270V0L
1
16
42
📣 Amazing News! We're happy to announce, that Brenda Schulman receives an ERC Advanced Grant for her project UPSmeetMet! Congratulations to all ERC-Awardees! 🎉 ➡️ more: https://t.co/STVQoC7BOC
@ERC_Research #ERCAdG #Ubiquitin
1
8
71
#Ubiquitin ligase substrate selectivity regulated kinetically by E3 self-degron mimicry. International effort by Schulman lab, @DanielS20127197 Scott & Moeko King & team @StJudeResearch @KheewoongBaek @MPI_Biochem & Gary Kleiger @unlv ➡️ https://t.co/T87thPUT4w
0
20
38