Alberto Valdeolivas
@alvaldeolivas
Followers
308
Following
146
Media
11
Statuses
433
Scientist in #bioinformatics @Roche. Physicist, Biological networks, System Biology, Spatial Omics, Cancer
Basel, Switzerland
Joined April 2016
Excited to share our new preprint! We map the spatiotemporal dynamics of therapy response & minimal residual disease in BRCA1;p53-deficient breast #cancer using #MultiOmics. We identify drug-tolerant EMT cancer cells, offering new therapeutic targets. 🔗 https://t.co/VostKuvNOw
0
3
2
Take a look inside a SARS-CoV-2-infected mouse lung with X-Pression! Our 3D reconstructions reveal changes in gene expression programs across the entire organ, leveraging ST and micro-CT. Check out the details in our preprint: https://t.co/1QZS4kVaKW
#SpatialTranscriptomics
0
2
2
Excited to share our new #biorxiv preprint! X-Pression combines deep learning with micro-CT to reconstruct 3D #spatialtranscriptomics from a single 2D section. A powerful, cost-effective way to explore tissues in 3D! Link: https://t.co/1QZS4kVaKW
#SARSCoV2-infected lung below!
0
1
2
Check out our GitHub to try Chrysalis! https://t.co/VuzUEvsqla
github.com
Powerful and lightweight package to identify tissue compartments in spatial transcriptomics datasets. - rockdeme/chrysalis
0
0
1
Chrysalis can be easily integrated into any workflow based on #Scanpy and works seamlessly with multi-sample datasets. This amazing work was led by @Rockdeme. A huge thanks to Jelica Vasiljevic, Kerstin Hahn and @RottenbergSven for their invaluable contributions.
1
0
0
Leveraging deep learning, we integrated morphological features from H&E images to uncover tissue compartments defined by morphology and expression. We also tested Chrysalis on multiple platforms, such as #VisiumHD, #SlideSeq, and #StereoSeq.
1
0
0
We benchmarked Chrysalis against other matrix decomposition and spatial domain inference methods with a focus on the #Visium platform, using both in silico and real-world datasets, demonstrating superior performance.
1
0
0
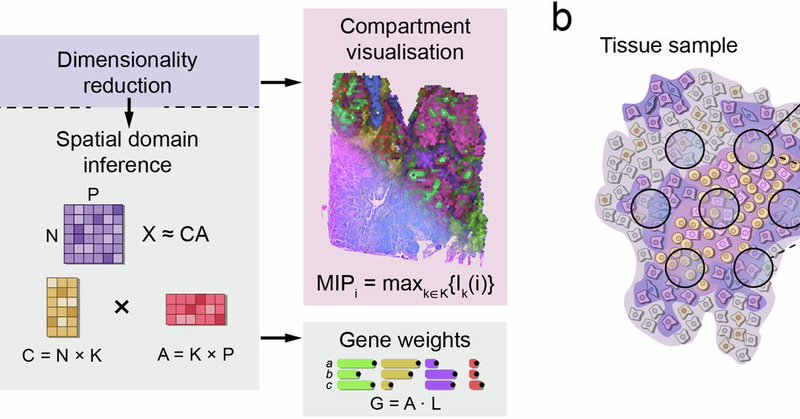
By fitting a simplex to the low-dimensional expression space, tissue compartments emerge as functionally distinct cellular niches.
1
0
0
Still using NMF? Try Chrysalis! Chrysalis combines spatially variable gene detection with archetypal analysis to decompose the gene expression matrix. The tissue compartments can be visualized simultaneously, with a unique twist.
1
0
2
Happy to announce that Chrysalis is finally out in @CommsBio ! Our new reference-free method identifies cellular niches in #spatialtranscriptomics with robust performance. https://t.co/2gpv1kkKHS
1
1
5
Super happy to announce that Chrysalis is finally out! Our new reference-free method identifies cellular niches in #spatialtranscriptomics with robust performance. Paper: https://t.co/XXPwC1Ze6D 🧵 (1/7)
1
1
2
Chrysalis, a machine learning-based framework, accurately infers cellular niches and underlying gene expression programs in the tissue from spatial transcriptomics data https://t.co/B9TJN0Hlb6
@Rockdeme @RottenbergSven @alvaldeolivas
nature.com
Communications Biology - Chrysalis, a machine learning-based framework accurately infers cellular niches and underlying gene expression programs in the tissue from spatial transcriptomics data. It...
0
3
5
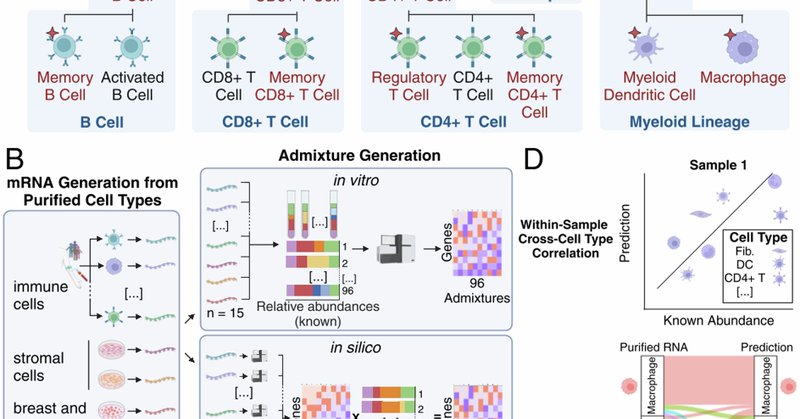
Paper is out! Deconvolve cellular composition from bulk gene expression
nature.com
Nature Communications - Deconvolution methods infer levels of immune infiltration from bulk expression of tumour samples. Here, authors assess 6 published and 22 community-contributed methods via a...
0
9
17
Check the latest @DR_E_A_M paper, evaluating methods to deconvolve cellular composition from bulk gene expression, with contributions from our (former) @alvaldeolivas & @JulioSaezRod 👇
0
7
26
New publication: The Molecular Landscape of Premature Aging Diseases Defined by Multilayer Network Exploration @cecilebeust @alvaldeolivas @AnthBaptista Galadriel Brière, Nicolas Lévy, @OzisikOzan @BaudotAnais
https://t.co/yZ0iigK7Zz
0
2
5
Hello! We're seeking an intern or master's student for a 6-month exploratory project in spatial analysis. Please share or apply! 😀 https://t.co/dCVXndvrb0
0
0
5
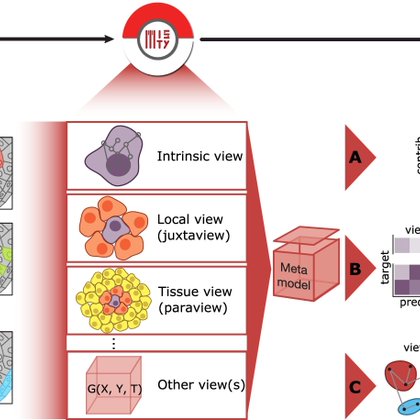
If you like MISTy https://t.co/ySyNNPW9eZ, you'll really like Kasumi https://t.co/00A3UjAcmf. Kasumi is a multi-view explainable representation learning approach, capturing localized relationship-based patterns in tissues across conditions aiming towards facilitating translation
genomebiology.biomedcentral.com
The advancement of highly multiplexed spatial technologies requires scalable methods that can leverage spatial information. We present MISTy, a flexible, scalable, and explainable machine learning...
Local heterogeneity of tissues can define their function and predict clinical outcomes. We introduce Kasumi 💻 https://t.co/2xbfYuo39I to identify spatially localized neighborhoods of intra and intercellular relationships from spatial omics persistent across samples & conditions
1
10
33
Finally out a new paper from the lab lead by @TrimbourR "Molecular mechanisms reconstruction from single-cell multi-omics data with HuMMuS" https://t.co/2DE1zzgfNG
#singlecell #multilayernetworks
0
23
85
Local heterogeneity of tissues can define their function and predict clinical outcomes. We introduce Kasumi 💻 https://t.co/2xbfYuo39I to identify spatially localized neighborhoods of intra and intercellular relationships from spatial omics persistent across samples & conditions
2
54
177
The molecular landscape of premature aging diseases defined by multilayer network exploration @cecilebeust @alvaldeolivas @AnthBaptista @OzisikOzan @BaudotAnais
https://t.co/udLkag0shs
0
1
4