Demeter Turos
@Rockdeme
Followers
27
Following
785
Media
14
Statuses
25
Doing Machine Learning and Spatial Omics @UniBern/Roche
Joined January 2012
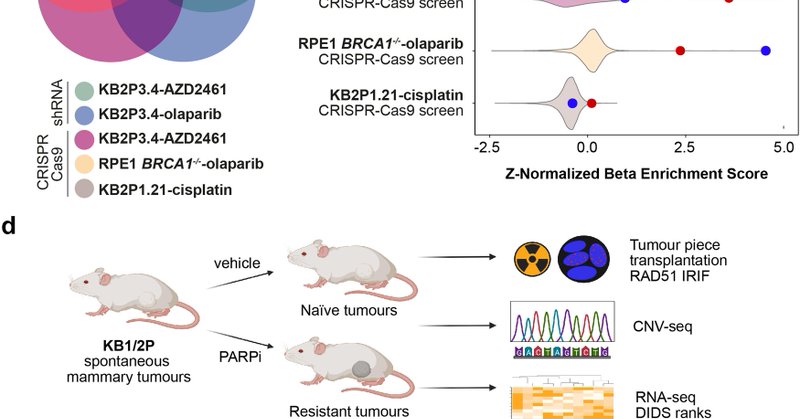
Excited to share our new preprint! We map the spatiotemporal dynamics of therapy response & minimal residual disease in BRCA1;p53-deficient breast #cancer using #MultiOmics. We identify drug-tolerant EMT cancer cells, offering new therapeutic targets. 🔗 https://t.co/VostKuvNOw
0
3
2
Take a look inside a SARS-CoV-2-infected mouse lung with X-Pression! Our 3D reconstructions reveal changes in gene expression programs across the entire organ, leveraging ST and micro-CT. Check out the details in our preprint: https://t.co/1QZS4kVaKW
#SpatialTranscriptomics
0
2
2
Excited to share our new #biorxiv preprint! X-Pression combines deep learning with micro-CT to reconstruct 3D #spatialtranscriptomics from a single 2D section. A powerful, cost-effective way to explore tissues in 3D! Link: https://t.co/1QZS4kVaKW
#SARSCoV2-infected lung below!
0
1
2
Check out our GitHub to try Chrysalis! https://t.co/YErV4zTXMj 🧵 (7/7)
github.com
Powerful and lightweight package to identify tissue compartments in spatial transcriptomics datasets. - rockdeme/chrysalis
0
0
0
Chrysalis can be easily integrated into any workflow based on #Scanpy and works seamlessly with multi-sample datasets. A huge thanks again to Jelica Vasiljevic, Kerstin Hahn, @RottenbergSven, and @alvaldeolivas for their invaluable contributions. 🧵 (6/7)
1
0
0
Leveraging deep learning, we integrated morphological features from H&E images to uncover tissue compartments defined by morphology and expression. We also tested Chrysalis on multiple platforms, such as #VisiumHD, #SlideSeq, and #StereoSeq. 🧵 (5/7)
1
0
0
By fitting a simplex to the low-dimensional expression space, tissue compartments emerge as functionally distinct cellular niches. 🧵 (3/7)
1
0
0
Still using NMF? Try Chrysalis! Chrysalis combines spatially variable gene detection with archetypal analysis to decompose the gene expression matrix. The tissue compartments can be visualised simultaneously, with a unique twist. 🧵 (2/7)
1
0
0
Super happy to announce that Chrysalis is finally out! Our new reference-free method identifies cellular niches in #spatialtranscriptomics with robust performance. Paper: https://t.co/XXPwC1Ze6D 🧵 (1/7)
1
1
2
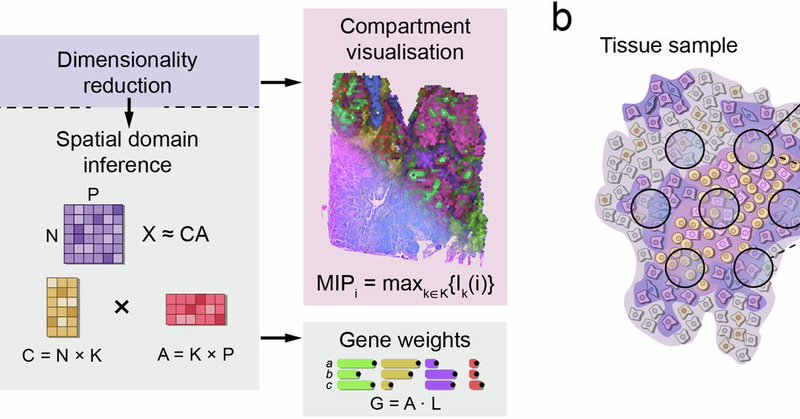
Chrysalis, a machine learning-based framework, accurately infers cellular niches and underlying gene expression programs in the tissue from spatial transcriptomics data https://t.co/B9TJN0Hlb6
@Rockdeme @RottenbergSven @alvaldeolivas
nature.com
Communications Biology - Chrysalis, a machine learning-based framework accurately infers cellular niches and underlying gene expression programs in the tissue from spatial transcriptomics data. It...
0
3
5
Happy to share our latest finding on H2AX promoting replication fork degradation and chemosensitivity in BRCA-deficient tumors https://t.co/q6Ydry042k
nature.com
Nature Communications - Histone H2AX has a known role in DNA damage repair but interestingly, its loss is associated with resistance to poly(ADP-ribose) polymerase (PARP) inhibition in...
2
10
48
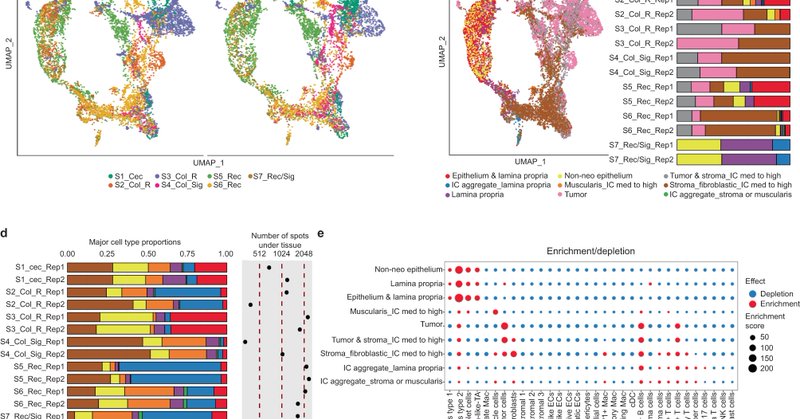
Excited to share that our paper “Profiling the heterogeneity of colorectal cancer consensus molecular subtypes using spatial transcriptomics” is now published in NPJ Precision Oncology! #CancerResearch #SpatialTranscriptomics #PrecisionOncology @Nature_NPJ
https://t.co/QgrLLzYR0D
nature.com
npj Precision Oncology - Profiling the heterogeneity of colorectal cancer consensus molecular subtypes using spatial transcriptomics
2
16
63
Chrysalis can be easily integrated into any workflow based on #scanpy and works on multi-sample datasets. Huge thanks to Jelica Vasiljevic, Kerstin Hahn, @RottenbergSven, and @alvaldeolivas for their invaluable contributions. 🧵 (5/6)
1
1
1
Leveraging deep learning, we integrate morphological features from the H&E image to discover unique tissue compartments. We also demonstrate the versatility of chrysalis using #Visium, #Slideseq, and #Stereoseq data. 🧵 (4/6)
1
1
3
We compare chrysalis with other matrix decomposition/spatial domain inference methods, like NSF and MEFISTO, relying on #cell2location and #Xenium reference data. 🧵 (3/6)
1
1
2
Chrysalis combines spatially variable gene detection with archetypal analysis to decompose the gene expression matrix, and employs MIP for simultaneous compartment visualization. 🧵 (2/6)
1
1
1
Super excited to share the preprint of chrysalis, our method that identifies tissue compartments in #spatialtranscriptomics without external reference. Preprint: https://t.co/WlJzMcj92k 🧵 (1/6)
2
5
8
Interested in a PhD project on radioresistance in Bern? Check this out:
0
10
10