Shuo Han
@Shuo_Han_
Followers
545
Following
384
Media
3
Statuses
89
Molecular labeling for biology and medicine. PI at Shanghai Institute of Biochemistry and Cell Biology.
Joined November 2020
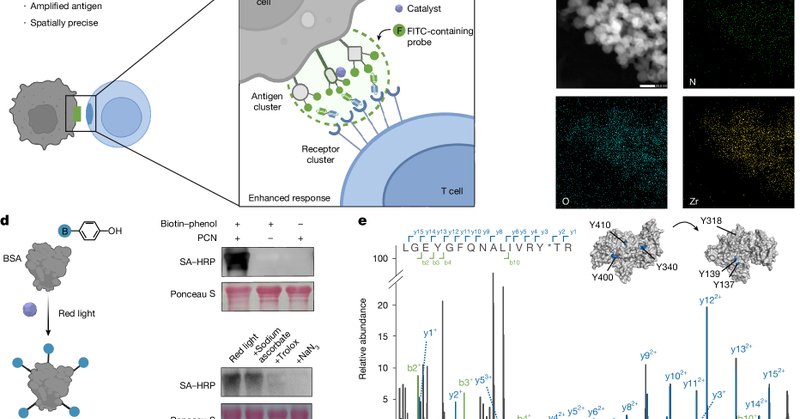
Excited to share the first paper from my lab in @Nature ! We repurposed the widely used research tool, proximity labeling, to demonstrate its therapeutic potential in antigen amplification for immunotherapy using mouse tumor models and patient samples. https://t.co/LISBT6GzXB
nature.com
Nature - Red light- or ultrasound-controlled proximity labelling is engineered to stimulate clustering-induced receptor activation and downstream signalling amplification to promote antitumour...
13
96
618
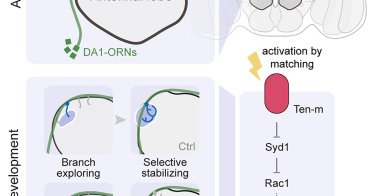
Huge congrats to @Raiseemhigh and all collaborators @BroadProteomics @aliceyting @Shuo_Han_ @StanfordBrain @HHMINEWS @HHMIJanelia! (Sneak in an ad: my lab at Janelia is seeking a highly motivated postdoc interested in kidney physiology. Pls share!) https://t.co/G6Z0hxol27
cell.com
Synaptic partner matching in the fly olfactory circuit is achieved by selectively stabilizing axon branches by partner dendrites. Synaptic partner matching molecule Ten-m regulates this process by...
0
8
37
Today, I would like to honor the memory of Roger Y. Tsien, who died on August 24, 2016. His legacy lives with all who use his technologies, including calcium sensors, fluorescent proteins, the acetoxymethyl (AM) ester, & many more! #FluorescenceFriday
https://t.co/YdWfBPUOBB
1
88
600
Lots of fun at our first lab outing and my first foray into poetry 😂
1
0
30
The FASEB BioAdvances journal team would like to welcome new Editorial Board member @Shuo_Han_ of the Shanghai Institute of Biochemistry and Cell Biology! Welcome aboard! https://t.co/qplmVSPvRW
#FASEBBioAdvances
1
1
3
Our work on ENTER is finally out @CellCellPress! Like📱that can 📷📞🎧, ENTER is a modular viral platform that unifies multiple applications: ⭐️Decode Ligand-Receptor interaction ⭐️Receptor-specific cargo delivery ⭐️Link receptor specificity with cell state at single cell
10
100
456
Woohoo! Big congrats to @JiaqiShen1, @LequnGeng and Xingyu Li for all their collaborative work leading to a brand new publication in @naturemethods! Our group teamed up with Prof Peng Li’s Lab 👏🏻!!! (@UMLifeSciences & @UMichDentistry)
4
7
66
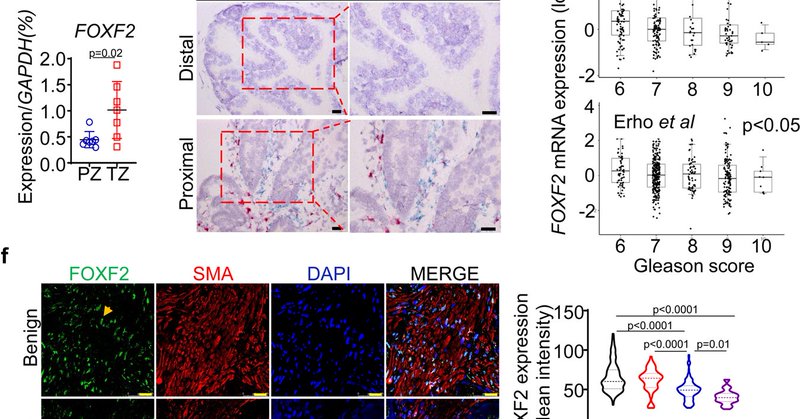
Prostate cancer occurs more frequently at the peripheral zone (PZ) than in transition zone (TZ) prostate, but the underlying mechanism is unclear. Study by Dr. Deyong Jia in my lab at @NatureComms provides one explanation for this clinical observation.
nature.com
Nature Communications - Forkhead transcription factor FoxF2 plays a crucial role in the development of organs derived from primitive gut. Here the authors show that reduction of Foxf2 expression in...
6
17
71
Thanks a ton to @DamonRunyon for organizing our amazing retreat! Fantastic talks from all fellows and really inspirational career advice from former alum @ValerieJHorsley. Also visited many friends and mentors at MIT Chem first time since I left 2016. See you next time Boston!
0
1
42
CRISPR can cut DNA and RNA, but did you ever wonder other activities might exist? Today in @biorxivpreprint, we show that the Type III-E Cas7-11 system programmably activates a nearby protease as part of microbial defense. + @jgooten @hnisimasu 🧵👇🏼 https://t.co/QIg548Jjy8 1/9
biorxiv.org
The type III-E Cas7-11 effector nuclease forms a complex with a CRISPR RNA (crRNA) and the putative caspase-like protease Csx29, catalyzes crRNA-guided target RNA cleavage, and has been used for RNA...
4
41
202
You know CRISPR complexes can cut DNA/RNA….what if some of them cleaved proteins too? Today in @biorxivpreprint, we (+ @omarabudayyeh @hnisimasu) continue our study of Cas7-11 and type III-E systems, revealing a new triggered protease function https://t.co/MJEm3OZx49 1/6
biorxiv.org
The type III-E Cas7-11 effector nuclease forms a complex with a CRISPR RNA (crRNA) and the putative caspase-like protease Csx29, catalyzes crRNA-guided target RNA cleavage, and has been used for RNA...
4
154
601
With more grants coming, we are excited to recruit postdocs who are interested in regenerative biology and extreme physiology. Some recent pubs and reviews here: https://t.co/K585rxxfSq,
https://t.co/5QaJp749SD,
https://t.co/3UG1FAk70m,
https://t.co/h8mJFgU6Dz Please RT.
4
64
172
I will start building my lab this summer in the brand new @westlake_uni, and we are hiring at all levels. Come join us to create protein-based synthetic intelligence: https://t.co/rpdhx9zULb
35
55
311
Highly recommend! Ken is a wonderful scientist and mentor! A fantastic choice if you are interested in applying cutting edge chemical biology tools to studying molecular physiology!
Excited to announce that the Loh lab will open in August @YaleWestCampus’s Institute for Biomolecular Design and Discovery! Our lab will use chemo-proteomic tools to study peripheral neurobiology in adipose tissue and the pancreas. We are hiring at all levels, please DM!
0
0
4
Thank you again to all of the attendees @epiSCniche_GRC! We are building a strong, inclusive, and welcoming community of exceptional #stemcell researchers. It was a privilege to organize this meeting together with @ValerieJHorsley, and I hope to see all of you again in two years.
0
3
36
As a beautiful sequel to @JiefuBiol & @Shuo_Han_’s 2020 Cell paper describing cell surface proteome profiling in vivo, this new paper by @chichi_xie & Liqun Luo et al. performs cell surface proximity labeling in WT vs. Acj6 knockdown flies, to discover... https://t.co/fvM8qELcqU
1
3
47
Congrats Jiefu! So happy for you! Look forward to all the great science from Li Lab and hopefully we find something to collaborate again in the near future haha!
Excited to share the news: I will start my lab @HHMIJanelia and join the 4D Cellular Physiology area. We will work on systems cell surface biology, combining method development and mechanistic investigation on neuro-immune crosstalk.
1
0
1
Thrilled to share our work on the mapping of RNA content of nuclear domains by APEX-seq! “Systematic mapping of nuclear domain-associated transcripts reveals speckles and lamina as hubs of functionally distinct retained introns” is out @MolecularCell
https://t.co/MGvYB8xaqx (1/8)
13
64
253
Come join our team!
Broad Proteomics is looking for a Research Associate interested in proteomics research. Job desc: https://t.co/tWmxE3ffJB . Please send your applications to jobs-proteomics@broadinstitute.org. @HasmikKeshishi1 @broadinstitute #jobs
1
3
4