Rasim Barutcu
@rassobar
Followers
710
Following
1K
Media
22
Statuses
1K
Associate Director @ ScitoVation Transcriptomics, Toxicogenomics, Risk Assessment, Chromosome Biology, Epigenetics, RNA Biology
Joined February 2016
Great work! Congrats Sheq!
It's my pleasure to present the next big preprint from SheqLab! An exciting application of our O-MAP platform that I hope will transform the study of nuclear architecture. If you've ever wanted to dissect the subnuclear "neighborhood" around an individual locus, read on! (1/30)
0
0
0
Glad to have contributed to this work! Great insights on inter-chromosomal interactions… 👇🏻👇🏻👇🏻
🚨Inter-chromosomal contacts (‘kissing chromosomes’) organize human genome topology along a spatial gradient of genome activity (62 Hi-C datasets). Check https://t.co/a5izMPo6C4,
@NatureComms, @4DNucleome, @The3DGenome, @humangenomeorg, @netscisociety, @Emblebi, @MoGen_Grads
1
1
8
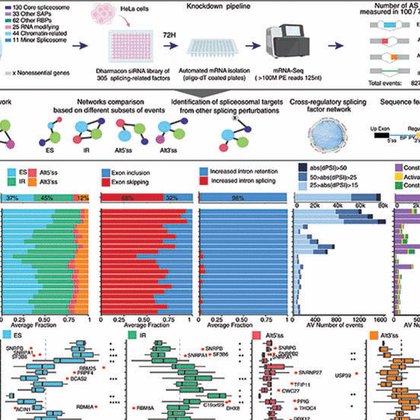
Introns? Splicing? Anyone? Take a sip.. or a gulp: https://t.co/84pWq3eYlu
science.org
The spliceosome is the complex molecular machinery that sequentially assembles on eukaryotic messenger RNA precursors to remove introns (pre-mRNA splicing), a physiologically regulated process...
0
0
3
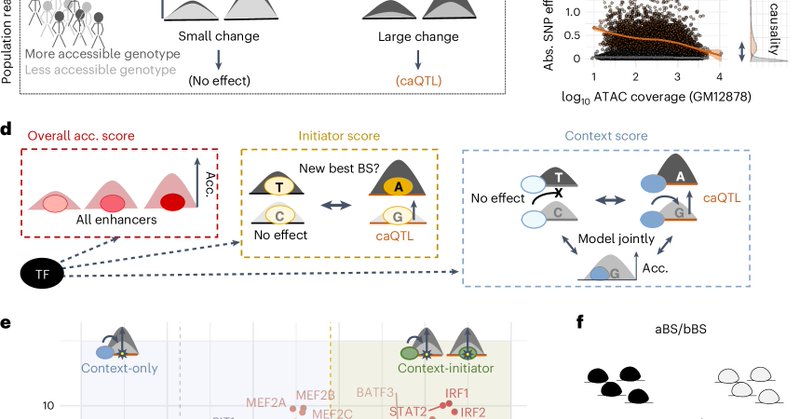
I haven't been tweeting for a while, but I think this is worth breaking the silence... Context transcription factors establish cooperative environments and mediate enhancer communication by @BartDeplancke and his team. https://t.co/NbWCfvNjKL
nature.com
Nature Genetics - This study identifies context-only transcription factors (TFs), a TF class that enhances DNA accessibility initiated by cell type-specific TFs and establishes cooperative...
0
0
5
What is happening?! Nuclear RNA forms an interconnected network of transcription-dependent and tunable microgels is happening...
biorxiv.org
The human cell nucleus is comprised of proteins, chromatin and RNA, yet how they interact to form supramolecular structures and drive key biological processes remains unknown. Conflicting models have...
0
0
2
mRNA secondary structure prediction using utility-scale quantum computers https://t.co/2dyUOV6Mad
arxiv.org
Recent advancements in quantum computing have opened new avenues for tackling long-standing complex combinatorial optimization problems that are intractable for classical computers. Predicting...
0
0
1
Horizontal transmission of functionally diverse transposons is a major source of new introns https://t.co/ou8ArSc1rF
#biorxiv_genomic
0
4
9
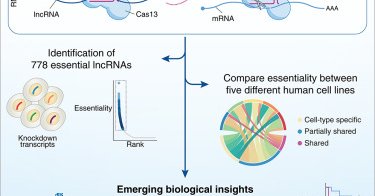
The "lncRNA" Malat1 is trafficked to the cytoplasm as a localized mRNA encoding a small peptide in neurons
pubmed.ncbi.nlm.nih.gov
Synaptic function in neurons is modulated by local translation of mRNAs that are transported to distal portions of axons and dendrites. The metastasis-associated lung adenocarcinoma transcript 1...
0
1
6
How do enhancers work over distances that sometimes exceed megabases? Excited to share our work led by @gracecbower where we uncover a unique sequence signature globally associated with long-range enhancer-promoter interactions in developing limb buds: https://t.co/n0p7BdxSdq 1/
23
232
693
Excited to share with the single-cell community the most recent work of @tmontsay and @AEsteveCodina from @cnag_eu, something we think will be a game-changer in #scRNAseq quality control: https://t.co/MeVrelcG9n
@biorxivpreprint Get ready for the tweetorial! 1/10
biorxiv.org
The advent of droplet-based single-cell RNA-sequencing (scRNA-seq) has dramatically increased data throughput, enabling the release of a diverse array of tissue cell atlases to the public. However,...
3
23
101
Our most recent preprint on #DeltaSplice for quantitative prediction of splicing and splicing-altering mutations. Great collaboration with Tao Jiang Lab. Led by Chencheng Xu@Jiang lab and a former postdoc @BaoSuying in my lab. Software available soon-stay tuned!
3
13
74
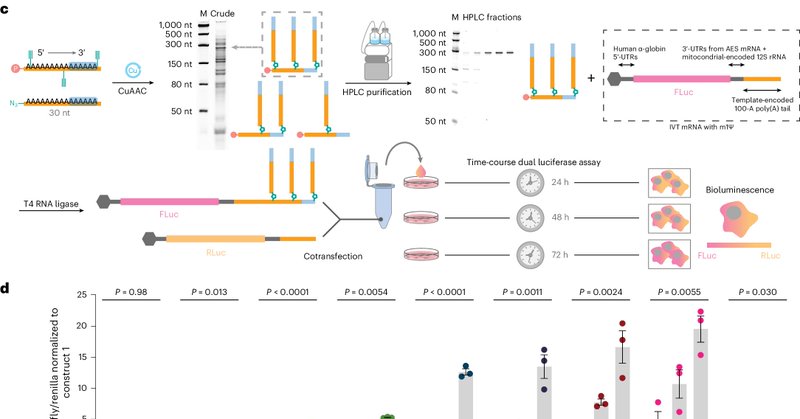
Branched chemically modified poly(A) tails enhance the translation capacity of mRNA https://t.co/2uvboExTRn
nature.com
Nature Biotechnology - mRNA with engineered poly(A) tails produces prolonged higher levels of protein.
0
0
0
Our paper describing Bioframe, a python library for working with genomic intervals, was just accepted!
academic.oup.com
AbstractMotivation. Genomic intervals are one of the most prevalent data structures in computational genome biology, and used to represent features ranging
Check out bioframe ( https://t.co/BGzBXFjJhB), a library for working with genomic interval #dataframes in #python and #pandas!!
2
13
44
https://t.co/NGBUL61109 I am very excited to share our recent paper on @Nature . It is a great collaboration with Dr. Murre and Dr. Fisch at UCSD. We discovered a molecular mechanism to shape nuclear morphology. @CCLRI
5
23
80
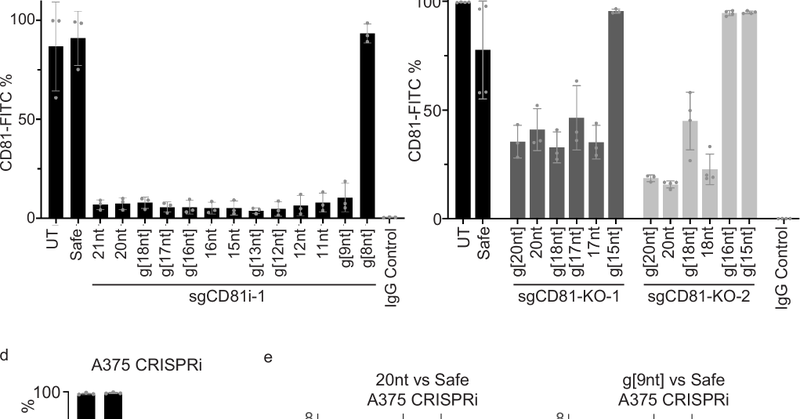
In an era of RNA delivery and therapeutics, we sought to “write” RNA molecules directly into messenger RNA to replace disease-causing exons or engineer synthetic proteins Our new work on RESPLICE enables fully programmable and highly efficient (45-90%) transcriptome engineering
9
100
592