Ben Lehner
@BenLehner
Followers
5K
Following
2K
Media
197
Statuses
3K
Head of Generative and Synthetic Genomics @sangerinstitute; Systems + Synthetic Biology @CRGenomica; Co-founder https://t.co/5N3PvqNXec https://t.co/l6NKYFgZBP
Barcelona | Cambridge
Joined April 2009
Allosteric and Energetic Remodeling by Protein Domain Extensions by @cristonah
https://t.co/gaGtxCj3Ju
biorxiv.org
Many functions of proteins are performed by independently folding structural units called domains. The structures of domains are conserved during evolution but they are not identical. For example,...
0
5
17
Under which circumstances do genomic neural networks learn motifs and their interactions? https://t.co/EfUyBQrjaX
biorxiv.org
The use of neural networks to model genomic data in sequence-to-function scenarios has soared over the last decade. There remains much debate about whether these models are interpretable, either...
3
17
90
🎲 Our paper on the genetics, energetics, and allostery in proteins with randomized cores and surfaces is out today @ScienceMagazine! 🧬 By charting a protein’s sequence universe, we could rationalize which versions were kept through evolution – and why many stable ones were not.
6
55
242
Only a few days left to apply to become a group leader in our Generative Biology programme at @sangerinstitute
https://t.co/g7sgCt53uK
sanger.ac.uk
We are a world-leading genomics research institute in Cambridge. Our work helps improve human health and understand life on Earth
0
19
18
New preprint: Allostery is a widespread cause of loss-of-function variant pathogenicity by the great @liao_xiaotian
https://t.co/i4T3JtEgVn.
biorxiv.org
Allosteric communication between non-contacting sites in proteins plays a fundamental role in biological regulation and drug action. While allosteric gain-of-function variants are known drivers of...
0
8
53
The evolution of allostery in a protein family. New preprint from the brilliant @ainamartiaranda. Seven complete allosteric maps.
1
9
46
Alzheimer’s disease 🧠starts with a molecular domino effect - but what triggers the first piece to fall? In our new study we cracked open the black box of early protein aggregation, and the findings could reshape how we fight neurodegeneration. 🧵👇 https://t.co/PQZcE4bGwN
science.org
The aggregation rates of >140,000 mutants reveal the energetic structure of an amyloid-β nucleation reaction transition state.
2
12
29
Come and join us! We’re hiring a new Group Leader in Generative Biology at the @sangerinstitute Building AI models or the data to train them? Core funding of >$130M a year for a faculty of ~30. pls RT! https://t.co/skA0MAOTyZ
https://t.co/cZ8i3TigJH
1
27
61
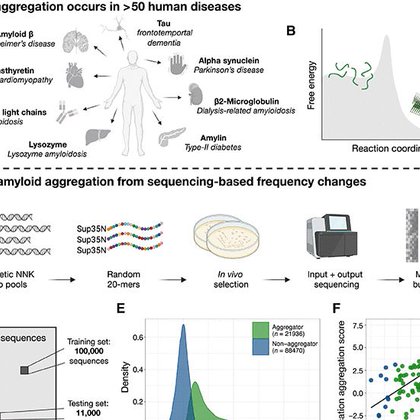
New publication. We quantify the aggregation of >100,000 random protein sequences to train CANYA, a convolution-attention hybrid neural network to predict aggregation from sequence. With @Bennibolo @CRGenomica @sangerinstitute @IBECBarcelona
science.org
xAI trained on >100,000 random peptides predicts and elucidates primary sequence determinants of aggregation.
2
38
151
I’m excited to co-host the new #AIxBio conference alongside @Avsecz, @ferruz_noelia, @pdhsu, @BenLehner, @ml19, @deboramarks, @p_tingying, @CarolineUhlerat at @wellcomegenome/ @sangerinstitute! We’ll focus on modellingz and designing DNA, RNA, proteins, and cells, as well as
3
52
198
Could #AI be a game-changer for #BiologicalResearch? Join us for #AIxBio25, our new conference on the transformative potential of #AI and #MachineLearning, in shaping the future of medical science. 📅Save the date: 23-25 June 2025 📎 Find out more: https://t.co/yONsRLQESs
0
14
28
3 great computational positions open at @ALLOXbio to work with the great @aj4re @JuliaDiumenge @pbcenturion @mseumaar
At @ALLOXbio we now have 3 computational positions available! 📌Structural Bioinformatics Scientist 👉 https://t.co/owf7tlAUWr 📌Platform Bioinformatician 👉 https://t.co/JwtjcOyoBi 📌Computational Chemist 👉 https://t.co/WkEQUFFf1M Apply or repost to help us spread the word 🙏
0
1
2
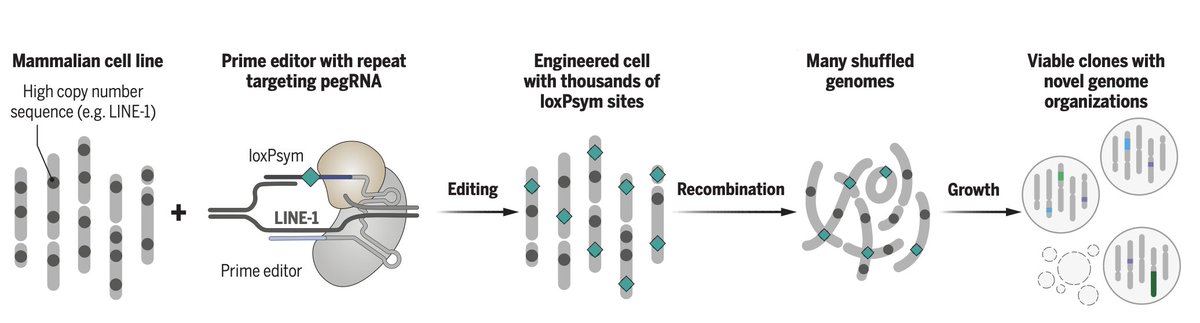
I’m delighted to share our work on scrambling the human genome using prime editing, repetitive elements, and recombinases @ScienceMagazine! @ferreira_raph_ @LeopoldParts @ProfTomEllis, @geochurch @SangerInstitute
https://t.co/UtJ13iNRAq Lots of changes from the preprint. A🧵
9
83
321
If you’d like to work in this exciting science area, @LeopoldParts lab at the @sangerinstitute is hiring postdocs! This is all part of the new Generative and Synthetic Genomics programme with nearby @benlehner and Jussi Taipale
0
5
16
We are happy to share our enhancer scramble story, a strategy to create hundreds of stochastic deletions, inversions, and duplications within mammalian gene regulatory regions and associate these new architectures with gene expression levels 🧵 @LeopoldParts, Pierre Murat
2
33
168
Introducing Human Domainome v1, the largest and most comprehensive library of human protein variants to date, which maps the effects of +500K mutations across 522 domains. The study by @BenLehner and Toni Beltran @mutatoni is out in @Nature. ✍️ Illustration by @QueraltTolosa
1
12
22
Protein instability is the main driver of inherited genetic conditions, according to the largest map of protein variants to date − Human Domainome 1.0🔎 This dataset can help predict how proteins behave, paving the way for new treatments💊 https://t.co/7lIRduqP64
3
29
129
Site-saturation mutagenesis of 500 human protein domains:
nature.com
Nature - Large-scale experimental analysis of Human Domainome 1, a library containing more than 500,000 missense mutation variants across more than 500 human protein domains, reveals that 60% of...
0
25
90
The first ‘human domainome’ reveals the cause of a multitude of diseases. An ingenious large-scale experiment in Barcelona (by @mutatoni & @BenLehner) has led to the creation of a catalog detailing the effects of half a million DNA mutations: https://t.co/oE9kfCX2nT
english.elpais.com
An ingenious large-scale experiment in Barcelona has led to the creation of a catalog detailing the effects of half a million DNA mutations
0
1
3