Albert Escobedo
@AlbertIltirda
Followers
278
Following
634
Media
29
Statuses
137
Postdoc @ the CRG Genetic Systems lab | Protein biophysics - Deep Mutational Scanning - NMR - Molecular Dynamics
Joined October 2011
🎲 Our paper on the genetics, energetics, and allostery in proteins with randomized cores and surfaces is out today @ScienceMagazine! 🧬 By charting a protein’s sequence universe, we could rationalize which versions were kept through evolution – and why many stable ones were not.
6
55
241
🙏 Huge thanks to my amazing co-authors Gesa Voigt & @aj4re for their key contributions, and to @BenLehner for his great guidance! 📝 Grateful to the reviewers for their thoughtful feedback, and everyone @ScienceMagazine for helping bring this work to light.
0
0
3
🔬 Our findings suggest models of protein evolution must account for energy couplings and allosteric constraints. 🚀 These insights could accelerate protein engineering—for example, guiding resurfacing to reduce immunogenicity via smarter directed evolution.
1
0
4
🧪 But stability isn’t the whole story—what about function? 🎯 Not all stable core variants could bind the FYN ligand. 🧿 Our findings suggest allostery is to blame: core mutations can subtly impact function—and become catastrophic when they pile up.
1
0
2
🧩 Many amino acid combinations from homologs cores worked when “transplanted” into FYN-SH3—but some didn’t. 🛠️ For the toughest cases, suppressor mutations outside the core rescued stability—thanks to energetic couplings across the protein.
1
0
1
🤖 We fed our large combinatorial mutagenesis datasets into an AI that trains fully interpretable energy models to predict protein variant stability. 🧮 These models accurately distinguished sequence combinations found in nature—from homologs that diverged billions of years ago.
1
0
1
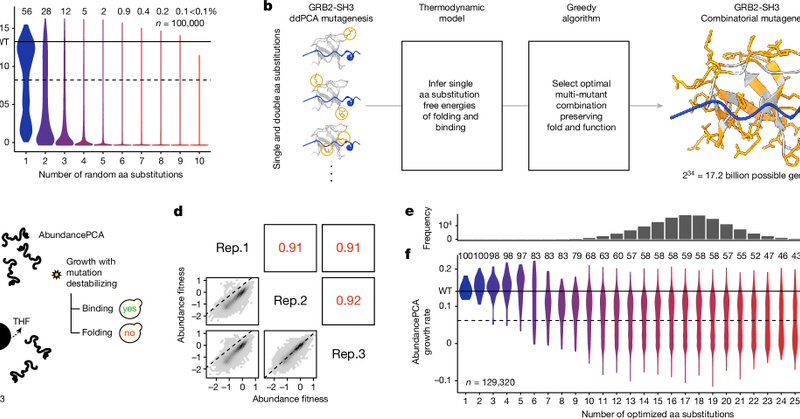
🎲 We randomized the core and surface of the human FYN kinase SH3 domain using reduced amino acid alphabets. 🎰 Thousands of amino acid combinations retained the domain’s stability. 🧱 Even load-bearing amino acids at the core were highly malleable.
1
0
3
🌌 A small protein has as many possible sequences as atoms are in the Universe: 10^78. 🔭 How can we explore and model such a vast universe of possibilities? 🦎 Does that help understanding how evolution found so many stable, functional sequences?
1
1
5
🤯 Mapping energy couplings in the Aβ42 nucleation transition state? 💪🏼 No problem for crack team @immunobananna @mseumaar @aj4re @BenLehner & @Bennibolo! 🧠 A massive tour-de-force in Alzheimer’s & neurodegeneration research — 📄 check out the paper and Anna’s awesome thread!
Alzheimer’s disease 🧠starts with a molecular domino effect - but what triggers the first piece to fall? In our new study we cracked open the black box of early protein aggregation, and the findings could reshape how we fight neurodegeneration. 🧵👇 https://t.co/PQZcE4bGwN
0
0
4
Thank you @Bennibolo @MafaldaFigDias @Jonnygfrazer for inviting me to share how we fit thermodynamic models to DMS data at this 8th MSS workshop 🙏🏼! It’s been a pleasure to share the floor with @ferruz_noelia and @NotinPascal and learn lots from them 👨🏻💻!
🔬🗣 Warming up for the 8th Mutational Scanning Symposium with a satellite workshop at #IBEC on how to choose a model for variant interpretation and protein design Great line-up of speakers for an interactive quantitative biology session!
0
2
7
I am incredibly excited to announce that our project with @BenLehner, @_ThomasWilhelm, and the @CRGenomica #TBDO has been awarded a Proof of Concept Grant from @ERC_Research! Huge thanks to everyone involved for their ongoing support – let's boost the science!
Congratulations to @benlehner @AlbertIltirda and @_ThomasWilhelm for receiving an @ERC_Research Proof of Concept Grant! The CRG team will explore the commercial potential of their research. #ERCPoC
https://t.co/sCJfZJBuze
2
0
11
GCAAGGACATATGGGCGAAGGAGA, las 24 letras cruciales en el surgimiento del autismo. Científicos del @IRBBarcelona han descubierto un mecanismo que podría explicar un elevado porcentaje de los trastornos del espectro autista: https://t.co/tVnxe9OmlO
elpais.com
Un equipo de científicos españoles descubre el mecanismo que podría explicar un elevado porcentaje de los trastornos del espectro autista
1
19
44
🧪 Are you passionate about molecular cloning? 🧬 Are you interested in applying ingenious methods for generating mutant libraries? 💊 Are you excited to join an exceptional team of scientists committed to revolutionising drug discovery? If so, here is a new job opportunity 👇
1
7
8
The genetic architecture of protein stability by @aj4re @ainamartiaranda @cristonah @ToniBeltran13 @JoernSchmiedel
https://t.co/Z4cxr9M4Hm
@CRGenomica @sangerinstitute @ALLOXbio
nature.com
Nature - By experimentally sampling from sequence spaces larger than 1010 and using thermodynamic models, the genetic structure of at least some proteins can be well described, indicating that...
2
70
241
Time to share my PhD work where we quantified the translational readthrough of thousands of pathogenic stop codons and trained predictive models to predict drug-induced readthrough of any stop codon. With the priceless guidance from @FranSupek +@BenLehner!
3
10
51
The simplicity of hydrophobic core fitness landscapes and pervasive allostery have important implications. Simple, interpretable models allowing accurate prediction over large evolutionary timescales should accelerate protein engineering and design and make it more understandable
0
0
0
To explore that, we fit three-state energy models of the folding and binding equilibria to the data. The inferred energy landscapes reveal that distal core mutations can have small cumulative effects on binding that can be catastrophic for function even if not too destabilizing.
1
0
1
Comparing the abundance and binding fitness for thousands of increasingly divergent variants reveals a surprising relationship: as the number of core mutations increases, the effects on binding appear greater than the effects on folding, suggesting pervasive core allostery.
1
0
2