Michael Vinyard
@vinyard_m
Followers
755
Following
3K
Media
26
Statuses
640
Deep neural networks and dynamical modeling of biology PhD @Harvard
Boston, MA
Joined March 2019
I am grateful for such a nice day. This was a great way to conclude this journey. Thank you!.
CONGRATULATIONS 🎉to the @getzlab and @lucapinello labs’ newest minted PhD, @vinyard_m, on successfully defending his thesis yesterday!! We are all so proud. @BroadInstitute @MGHCancerCenter @Harvard. 🥂🥂🥂
0
0
12
Thank you! I have been so lucky to have such great mentors.
Thrilled to celebrate @vinyard_m, a PhD student co-supervised by @gaddyg's lab, who successfully defended his thesis today. Congratulations, Dr. Vinyard! Special thanks to the examiners @brian_b_liau @JD_Buenrostro and everyone in the labs who contributed to this remarkable day!
7
0
35
RT @lauradmartens: Is binarization of scATAC-seq data really necessary? The conclusion from our analysis is that a quantitative treatment i….
0
97
0
RT @bloodgenes: Remarkable to see that our discoveries over 15 years ago would enable such tremendous advances for sickle cell disease and….
0
113
0
Congrats, team!.
Absolutely thrilled to share this incredible work led by @JayoungR ! A huge shoutout to the collaborative efforts with @SherwoodLab, @ccassa, their teams and several key collaborators. Check out her excellent twettorial for a summary and link to the preprint!#CRISPR #BEAN 🧬🌟☕️.
1
0
4
RT @GeffenYifat: So exciting to see these papers now published, an incredible team work. A huge thank you to our collaborators and co autho….
0
6
0
RT @nikos_trasan: 💥 I am thrilled to announce that Dictys (<δίκτυον), our tool to reconstruct dynamic GRNs from single cell multiomics data….
0
5
0
RT @nikos_trasan: Amazing talk by Luca Pinello on the new CRISPR-CLEAR tool! #Regsys #ISMBECCB2023 Conference, @ISCB #pinellolab #CRISPR h….
0
1
0
RT @lucapinello: 1/6 A little thread to celebrate this milestone! Our SIMBA manuscript has finally seen the light of day, after a journey o….
0
41
0
A tremendous privilege to be able to work with and learn from this team.
I am beyond excited to share that SIMBA, a versatile single-cell graph embedding method, has been published @naturemethods (1/6).
0
0
6
RT @velvetatom: Running an AI x BIO HACKATHON this month with @openbioml @huggingface @LatchBio. 🧬💻 - remote, GPU's & API's provided. Hel….
0
122
0
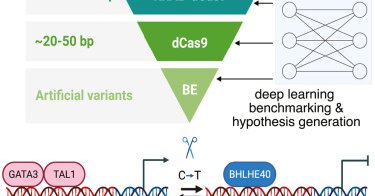
RT @ZeyuCHEN19: Excited to share our latest work dissecting a CD69 enhancer at base resolution out in @cellgenomics today .
cell.com
Chen and Javed et al. integrated epigenetic perturbations, base editing, and deep learning to dissect a CD69 enhancer. They identified a single C-to-T mutation that impacts gene expression by...
0
37
0
RT @getz_lab: Michael Vinyard @vinyard_m | Modeling single-cell dynamics using stochastic generative models based on neural differential eq….
0
4
0
If you’re not at AACR but you’re interested in single-cell dynamics, visit for early access to our Python package, scDiffEq, and to indicate your interest in collaborating. #AACR #singlecell #python #pytorch #GenerativeModels #ML #DeepLearning.
0
1
2