Jayoung Ryu
@JayoungR
Followers
265
Following
599
Media
11
Statuses
74
PhD Candidate at @BIG_Harvard_PhD with @lucapinello | Bayesian modeling/ML on perturbation & single cell data
Cambridge, MA
Joined September 2019
Absolutely thrilled to see the first work that I led during my PhD is finally out in @NatureGenet !! Check out the manuscript & Tweetorial 👇
We are very happy to see that our manuscript, 'Joint Genotypic and Phenotypic Outcome Modeling Improves Base Editing Variant Effect Quantification,' is now published in @NatureGenet. Read it for free here: https://t.co/Q2fu463hFD, original twittorial below!
4
4
34
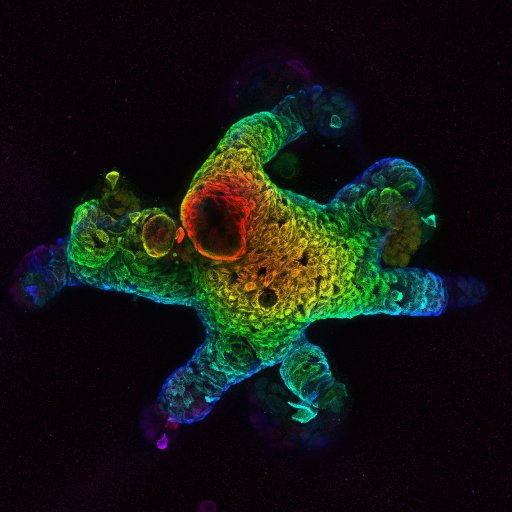
First pre-print from the lab! Together with @lucapinello and @danielevanbauer we developed CRISPR-CLEAR, an experimental and computational pipeline to dissect enhancers at nucleotide resolution. https://t.co/ZfiW9Dm4UG
5
54
265
Excited to share our work "CRISPR-CLEAR", an end-to-end workflow for dissecting enhancers at nucleotide resolution! Check out our pre-print and @DSeruggia's tweetorial below! Lead with @SWittibschlager and @zafateniac and labs @lucapinello @DSeruggia and @danielevanbauer.
First pre-print from the lab! Together with @lucapinello and @danielevanbauer we developed CRISPR-CLEAR, an experimental and computational pipeline to dissect enhancers at nucleotide resolution. https://t.co/ZfiW9Dm4UG
0
4
22
Congratulations to @JayoungR on winning the Best Paper Award at the ICML workshop on @AI_for_Science for her internship work with us at Genentech! We will also be presenting this work next month as part of an oral presentation at MLCB in Vancouver!
How can we integrate two single-cell perturbation screens like Perturb-seq and optical pooled screens? Thrilled to introduce Perturb-OT for cross-modality matching and prediction of perturbation responses! Work with amazing @_romain_lopez_ @_bunnech & Aviv (1/)
1
9
50
Excited to share our new paper on Contextual AI models for context-specific prediction in biology in @NatureMethods led by stellar @_michellemli
https://t.co/6cP5CKn9Xx Understanding how proteins work and developing new therapies requires knowing which cell types proteins act
7
60
287
🚀Excited to announce that I am joining @modella_ai as the Founding CTO! With @AI4Pathology @MYLu97 throughout my PhD, we have released several amazing foundation models (UNI, CONCH, and most recently PathChat published in Nature: https://t.co/IEWQXdUWCO). Now, we are going all
nature.com
Nature - PathChat, a multimodal generative AI copilot for human pathology, has been trained on a large dataset of visual-language instructions to interactively assist users with diverse pathology...
🚀We’re thrilled to come out of stealth and announce PathChat 2, the 1st multimodal generative AI copilot for pathology. PathChat 2 improves upon PathChat 1 (recently published @Nature, https://t.co/CK8MBYULOG).
https://t.co/yuSBTCUXwA Waitlist: https://t.co/NoeEnQnYQL 🧵1/2
11
7
83
@MLGenX thank you for organizing a fantastic workshop at @iclr_conf and for selecting our paper on DNA-Diffusion for the Outstanding Paper Award! Congratulations to the entire team that made this possible! #ICLR2024 🎉
1
4
27
This work opens a way to unify causal models of cell biology. Check out the full details below! 📰 https://t.co/vcew4G3yQP 💿 https://t.co/Ur9TRXROjo (5/5)
github.com
Cross-modality matching and prediction with labeled Gromov-Wasserstein Optimal Transport. - Genentech/Perturb-OT
0
5
24
We show that labeled GWOT methods lead to better 1) matching and 2) prediction compared to OT, GWOT, and adversarial VAE methods in Perturb-CITE-seq-like data. We also show that we can learn a better 3) feature-to-feature matching from better cell-to-cell matching. (4/)
1
0
1
For cross-modality matching, we modified GWOT methods to admit cells’ perturbation labels (“Labeled GWOT/COOT”) and show it can be sped up by # perturbations. It uses cells’ perturbation labels and the global topology of cellular responses for better matching. (3/)
1
0
1
We predict perturbation responses of unobserved (ex. RNA) from the observed (ex. cell morphology) modality by 1) learning cross-modality matching with labeled Gromov-Wasserstein Optimal Transport (GWOT), 2) then training predictor using the learned matching. (2/)
1
0
2
How can we integrate two single-cell perturbation screens like Perturb-seq and optical pooled screens? Thrilled to introduce Perturb-OT for cross-modality matching and prediction of perturbation responses! Work with amazing @_romain_lopez_ @_bunnech & Aviv (1/)
3
31
167
Thanks so much for the shoutout! 🤗 Happy to help and excited that BEAN is contributing to cool discoveries to come!
I want to give a huge shoutout to @JayoungR for being so kind and responsive in troubleshooting some issues I was having installing and running BEAN. All solved in 1 day! Also not the first time I've had a fantastic interaction with @lucapinello lab. Thanks for all of your help!
0
0
10
1/🧵Introducing Perturb-seq-in-the-loop: a sequential experimental design strategy for perturbation screens guided by multimodal priors, with 3X speedup over state-of-the-art active learning methods! With amazing @_romain_lopez_ @jchuetter Taka Kudo @antonio_science Aviv Regev
7
112
456
The SPECTRA package generates a series of splits with decreasing train-test similarity. Evaluating your models on these splits will give a better understanding of model generalizability. Read the preprint for more info on how this works: https://t.co/cgzHJaXttS
biorxiv.org
Deep learning has made rapid advances in modeling molecular sequencing data. Despite achieving high performance on benchmarks, it remains unclear to what extent deep learning models learn general...
1
2
9
Very excited to share our study on the benchmarking of methods to detect spatially variable genes (SVGs) and peaks (SVPs), a collaborative work with @zafateniac, @SongDongyuan, @GuanaoYan, @jsb_ucla, and @lucapinello. 🧵 (1/n)
1
13
47
Thrilled to share our inaugural work on spatial transcriptomics where we benchmarked methods for detecting spatially variable genes and peaks. Kudos to the team @ZhijianLi3,@zafateniac, @SongDongyuan, @GuanaoYan, @jsb_ucla for this great collaboration! https://t.co/MBzMDpZgRj.
biorxiv.org
Spatially resolved transcriptomics offers unprecedented insight by enabling the profiling of gene expression within the intact spatial context of cells, effectively adding a new and essential...
Very excited to share our study on the benchmarking of methods to detect spatially variable genes (SVGs) and peaks (SVPs), a collaborative work with @zafateniac, @SongDongyuan, @GuanaoYan, @jsb_ucla, and @lucapinello. 🧵 (1/n)
1
11
63