Pedro Ferreira
@pfalef
Followers
205
Following
3K
Media
1
Statuses
269
PhD in Computational Biology @cbg_ethz
Basel, Switzerland
Joined September 2018
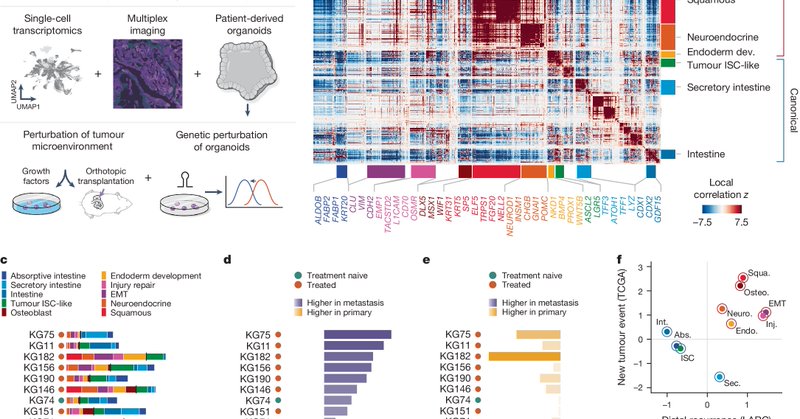
Excited to share one of our apex studies from our @NCIHTAN center, focused on the transition to metastasis, shedding light transition to #CRC #metastasis. Joint work with @KarunaMDPhD, led by @andrewrmoorman and @elliebenitez11. --> https://t.co/9mVfnoJtNe
nature.com
Nature - Colorectal cancer metastasis involves dramatic plasticity and loss of PROX1-mediated repression of non-intestinal lineages.
7
37
241
Congratulations on leading this amazing project @susanavinga!
This week all members were together in Basel, @ETH_en, for the final meeting of this amazing project. That's a wrap of these 3,5 years of training, exchanging, sharing & collaborating! Thanks to all the students, researchers and research administrators who have crossed our path!
1
0
2
This week all members were together in Basel, @ETH_en, for the final meeting of this amazing project. That's a wrap of these 3,5 years of training, exchanging, sharing & collaborating! Thanks to all the students, researchers and research administrators who have crossed our path!
0
1
9
Sigh. The level of profound confusion in the scRNA-seq analysis field keeps surprising me. I've now run into people that, in a discussion about technical artifacts of scRNA-seq, where I claimed there is no evidence for zero inflation, first insist that it does exist, and then 1/n
@NimwegenLab @maelouisewoods @GorinGennady @arjunrajlab The existence of ZI as a "technical artifact" is distinct from divergence from a NB or Poisson dist. This paper https://t.co/aMgAk4pk9g e.g. argues that zero inflation is observed for certain genes & is of Biological origin (& recommends still using a GLM with NB counts w/o ZI).
2
12
93
We have made many updates to our SCICoNE results. Everything is described in this new version of our preprint:
biorxiv.org
Copy number alterations are driving forces of tumour development and the emergence of intra-tumour heterogeneity. A comprehensive picture of these genomic aberrations is therefore essential for the...
0
3
3
The choice of whether to use Seurat or Scanpy for single-cell RNA-seq analysis typically comes down to a preference of R vs. Python. But do they produce the same results? In https://t.co/rVOiR847CY w/ @Josephmrich et al. we take a close look. The results are 👀 1/🧵
16
374
1K
I see we are getting to the stage of the discussion where people are starting to defend UMAP saying it can 'reveal patterns' or 'structure' in the data. Without ever specifying what precisely these patterns/structures represent. This is not surprising because hardly anybody 1/n
Thoughtful, well-informed counterpoint on the use of UMAP for visualising human genetic data, following the intense debate about the All Of Us paper this week.
4
118
515
Our latest out today in @Nature. We profiled 12 million single cells from mouse embryos spanning gastrulation to birth, defined cell type tree from zygote to birth, and unexpectedly found crazy fast changes within first hour of extrauterine life. OA PDF:
Excited to share our lab's latest preprint, led by @CXchengxiangQIU, @bethkarenmartin & Ian Welsh of @jacksonlab. We set out to build a single cell roadmap for all of mouse prenatal development, from single cell zygote to free-living pup. Preprint: https://t.co/GDS65Qf58X 1/n
15
153
580
An interesting approach to hierarchical cell type annotation for single-cell RNA-seq using a multi-level Bayesian factorization model.
biorxiv.org
Single-cell gene expression data characterizes the complex heterogeneity of living systems. Tissues are composed of various cells with diverse cell states driven by different sets of genes. Cell...
1
20
134
I really dislike this race to the bottom: No need to write protocols. No need to write your own introduction section. Just copy somebody else's. Everything is too much apparently. There are many reasons why being forced to formulate a protocol each time is helpful. 1. It 1/n
The arguments presented here against reuse of language in methods section are unconvincing. Yes, it is certainly possible to rewrite a protocol over and over again. Is that a valuable exercise in scientific writing ala Flaubert? No way. We're not writing Madame Bovary here.
2
4
21
We have made many updates to our scDEF method and results. Everything is described in this new version of our preprint: https://t.co/yVXkL5Ut4M
biorxiv.org
Single-cell gene expression data characterizes the complex heterogeneity of living systems. Tissues are composed of various cells with diverse cell states driven by different sets of genes. Cell...
0
0
5
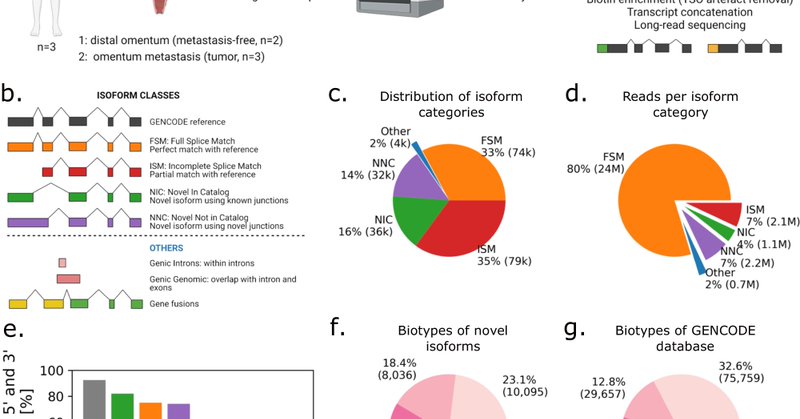
Delighted to present our full-length scRNA-seq study on ovarian cancer patients, in collaboration with the University Hospital Basel @labfranz ! https://t.co/I7LnLDd2gr Can we simultaneously detect isoform-level gene expression, mutations and gene fusions in cancer cells? 1/
nature.com
Nature Communications - Long-read single-cell RNA sequencing is capable of detecting isoform-level gene expression and genomic alterations such as mutations and gene fusions, thereby providing...
3
12
39
New data arrived for #BA286 / #Pirola in #Switzerland: the variant is observed at low abundance in multiple locations through our #Wastewater surveillance of #SARSCoV2 variants.
4
54
131
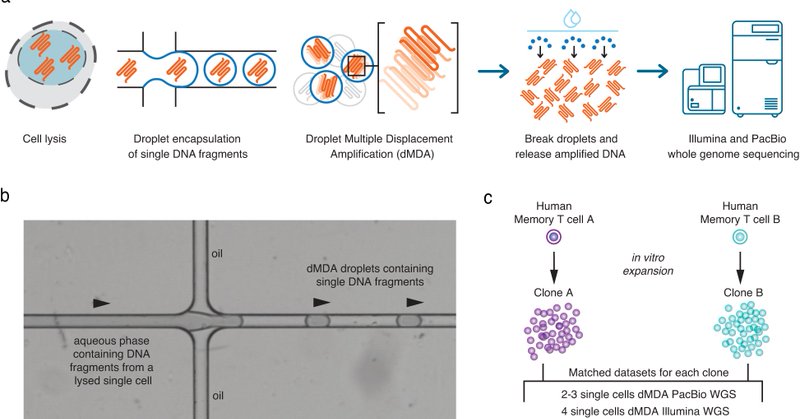
Thrilled to see our paper out today where we use long-read whole-genome sequencing to study SNVs, structural variation and repeats in single cells https://t.co/Qd05jzC9DG Many thanks to everyone involved @_adameur, @Jeff_Mold, @jakmic2003, @ngisweden, @scilifelab
nature.com
Nature Communications - Here the authors introduce a new method to study DNA in single cells by long-read sequencing. Their method gives a more complete view of the genomic structure of individual...
0
9
38
In our detection of BA.2.86 in two wastewater samples from Laupen (BE, Switzerland) of August 5 and 6: we observed co-occurrence of these mutations on the same fragments: – 21618: T, 21622: T, 21624: C, 21633: '---------', 21711: T, 21765: '------' (amplicon 67)
2
21
60
The next Computational Reproducibility seminar is this Wednesday at 5 pm: Kim Philipp Jablonski @kpj_py at @Google is going to speak about sustainable tool benchmarking and workflow development in Computational Biology. -- Join us for free at: https://t.co/eJc5KgJCQn
@SwissRN
0
3
2
We are pleased to welcome Prof Niko Beerenwinkel of @cbg_ethz. He joins the Institute as a @royalsociety Wolfson Visiting Fellow. He will work with @markowetzlab over the next two years to develop his research project "Single-cell copy number signatures as cancer #biomarkers".
0
1
11
🎙️ 1º episódio disponível - 𝗖𝟰𝗻𝗰𝗿𝗼 é o novo podcast do iMM-Laço Hub. A 1ª conversa @MCarmoFonseca @immolecular e Tamara Milagre, presidente @evitacancro, tem como mote “Como nasce um cancro na mama?” Spotify, Apple Podcasts e YouTube 🔗 https://t.co/I8k9PLPv2P
1
7
18
NOW OUT @ScienceMagazine 🧬🐾🤓 “The evolution of two transmissible cancers in Tasmanian devils” A 🧵 on our deep DNA sequencing dive into the startling genetic history of contagious tumours (1/n) 👇 https://t.co/XuV9LctJ4f
24
211
859