CBG, ETH Zurich
@cbg_ethz
Followers
913
Following
13
Media
15
Statuses
154
Computational Biology Group in D-BSSE, @ETH_en, led by Niko Beerenwinkel.
Basel, Switzerland
Joined September 2018
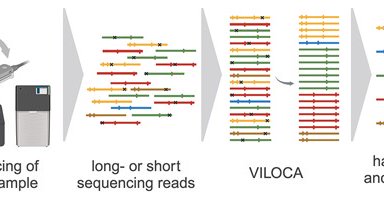
We are excited to share that our new tool VILOCA has been published in NAR G&B: .VILOCA is a novel method for mutation calling and local haplotype reconstruction from diverse viral sequencing data, which is also adept for viral surveillance in wastewater.
academic.oup.com
Abstract. RNA viruses exist as large heterogeneous populations within their host. The structure and diversity of virus populations affects disease progress
0
6
8
We're thrilled to announce the release of V-pipe 3.0! This computational pipeline is specifically crafted for analyzing short viral genomes using next-generation sequencing data. V-pipe 3.0 enables sustainable viral genomic data science. Check it out at:
academic.oup.com
Abstract. The large amount and diversity of viral genomic datasets generated by next-generation sequencing technologies poses a set of challenges for compu
0
1
3
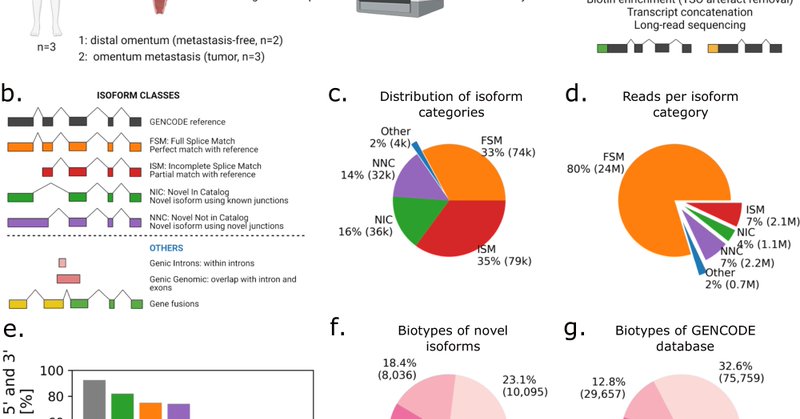
RT @ArthurDondi: Delighted to present our full-length scRNA-seq study on ovarian cancer patients, in collaboration with the University Hosp….
nature.com
Nature Communications - Long-read single-cell RNA sequencing is capable of detecting isoform-level gene expression and genomic alterations such as mutations and gene fusions, thereby providing...
0
13
0
Results obtained within our WISE (Wastewater Infectious disease Surveillance and Epidemiology) consortium @EawagResearch @ETH_BSSE supported by @snsf_ch and @BAG_OFSP_UFSP. @NEXUS_PHT @FGCZ_EN @TanjaStadler_CH @real_dr_david @DoktorYak @trj2 @CatharineAquino.
0
1
4
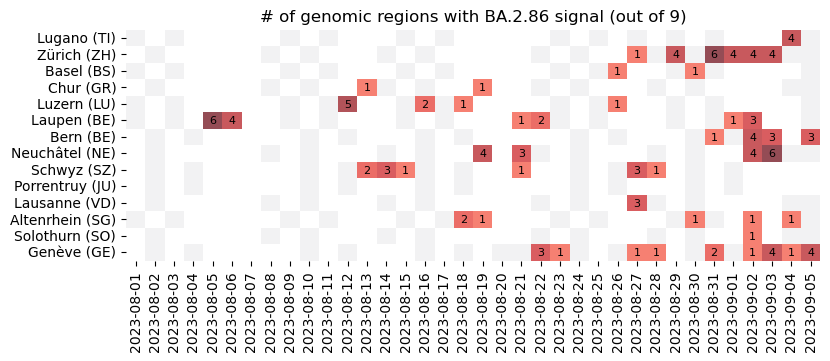
The analysis was performed using V-Pipe and COJAC. More details on our project and procedure available here and here
github.com
Procedure used for the surveillance of SARS-CoV-2, RSV and Influenza genomic variants in wastewater. - cbg-ethz/cowwid
1
0
3
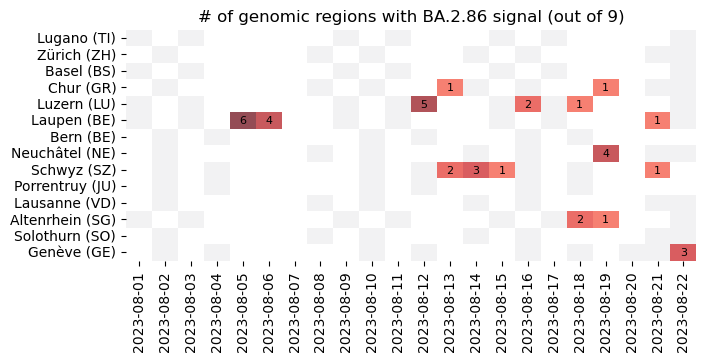
New batch of data analyzed, with further detections of #BA286 / #Pirola in multiple locations in #Switzerland through our #Wastewater surveillance of #SARSCoV2 variants. The BA.2.86 variant was detected in most of the treatment plants we survey.
2
17
33
Results obtained within our WISE (Wastewater Infectious disease Surveillance and Epidemiology) consortium @EawagResearch @ETH_BSSE supported by @snsf_ch and @BAG_OFSP_UFSP. @NEXUS_PHT @FGCZ_EN @TanjaStadler_CH @real_dr_david @DoktorYak @trj2 @CatharineAquino.
0
2
10
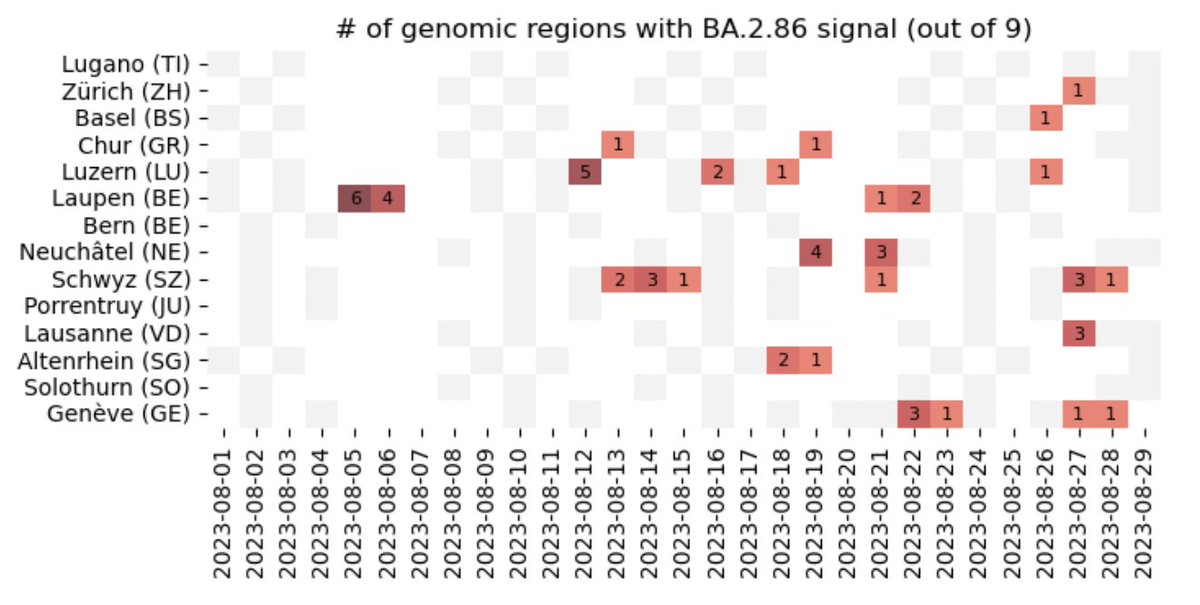
The analysis was performed using V-Pipe and COJAC. More details on our project and procedure available here and here
github.com
Procedure used for the surveillance of SARS-CoV-2, RSV and Influenza genomic variants in wastewater. - cbg-ethz/cowwid
1
1
7
New batch of data analyzed, with further detections of #BA286 / #Pirola in multiple locations in #Switzerland through our #Wastewater surveillance of #SARSCoV2 variants.
5
17
33
Results obtained within our WISE (Wastewater Infectious disease Surveillance and Epidemiology) consortium @EawagResearch @ETH_BSSE supported by @snsf_ch and @BAG_OFSP_UFSP. @NEXUS_PHT @FGCZ_EN @TanjaStadler_CH @real_dr_david @DoktorYak @trj2 @CatharineAquino.
0
1
4
The analysis was performed using V-Pipe and COJAC. More details on our project and procedure available here and here
github.com
Procedure used for the surveillance of SARS-CoV-2, RSV and Influenza genomic variants in wastewater. - cbg-ethz/cowwid
2
1
11
New batch of data analyzed, with further detections of #BA286 / #Pirola in multiple locations in #Switzerland through our #Wastewater surveillance of #SARSCoV2 variants.
4
19
48