Pedro Beltrao
@pedrobeltrao
Followers
7,038
Following
1,224
Media
370
Statuses
8,352
Associate professor at ETH ( @IMSB_ETH ) studying the cellular consequences of genetic variation. Part of @theLoopZurich .
Zurich, Switzerland
Joined March 2007
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
Merchan
• 317750 Tweets
National Anthem
• 128422 Tweets
Christian
• 127768 Tweets
Trump is Guilty
• 80746 Tweets
Farage
• 79112 Tweets
Osaka
• 65247 Tweets
The IEC
• 64649 Tweets
超・獣神祭
• 60516 Tweets
Bernabéu
• 45841 Tweets
Vargas
• 35732 Tweets
Fiorentina
• 35724 Tweets
Swiatek
• 27500 Tweets
Conference League
• 26258 Tweets
Mother Teresa
• 24949 Tweets
Eruviel
• 18880 Tweets
ナイトウェア
• 18682 Tweets
あなたの悪夢
• 17695 Tweets
$DAVIDO
• 13677 Tweets
Jerry Seinfeld
• 11721 Tweets
Amine
• 11521 Tweets
Last Seen Profiles
After 9 years of
@embl
, I am thrilled to say that next Jan I will join the faculty of
@ETH_DBIOL
at ETH Zurich with research at
@IMSB_ETH
. I am really excited about the move, my colleagues at IMSB and more broadly the amazing local research environment
68
17
494
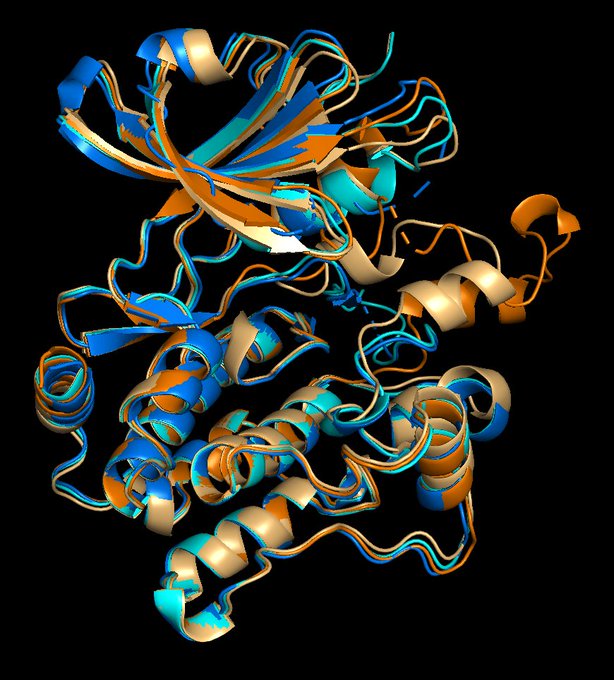
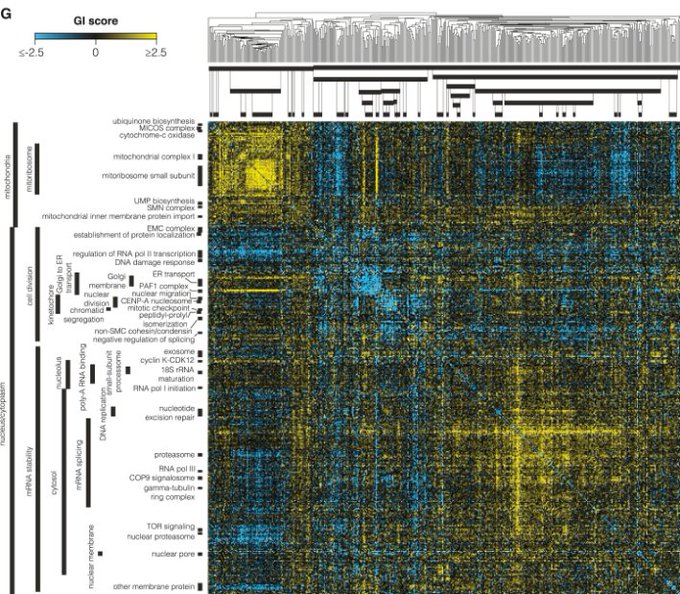
We joined a large community effort to assess diverse applications of AlphaFold 2 in the context of novel structural elements; missense variants; function and ligand binding sites; modelling of interactions and experimental structural data. Some highlights below:
2
153
401
Our collaboration with
@arneelof
's lab exploring the use of AlphaFold2 for large scale prediction of structures of human complexes is now published
@davidfburke
@Patrick18287926
et al.
9
101
362
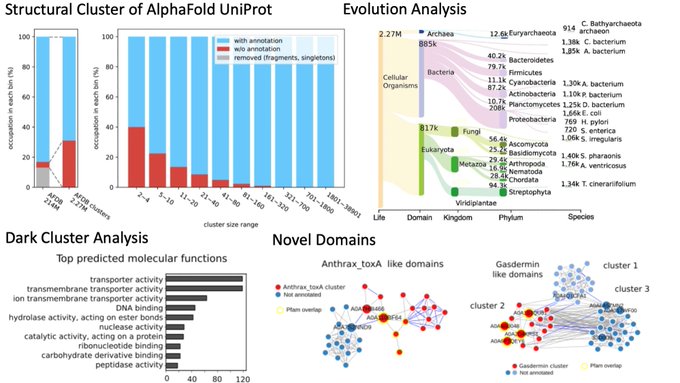
How many different protein shapes are there in 200 million predicted AlphaFold structures? In a joint project with
@thesteinegger
we (
@ibarrioh
,
@jgyyy15
et al) clustered these into around 2 million groups to study their novelty and evolution
8
111
344
Jointly with
@arneelof
's group we applied Alphafold to predict complex structures for 65,000 human protein-protein interactions. Here is what we learned in trying to generate a structurally resolved human interaction network
7
104
343
Less than 5% of human phosphosites have a known function. Also published today -
@d0choa
et al. have set out to create a high quality catalog of human phosphosites and an integrated score than can rank phosphosites critical for the cell
@NatureBiotech
6
136
294
Yesterday's
@emblebi
training webinar on "How to interpret AlphaFold structures" is now online. It is oriented towards users with practical explanations about the method, the confidence scores, the database and several use case applications
0
125
293
How does SARS-CoV-2 take over kinase signalling to subvert normal cell function and what drugs can we use to counter these changes? Our new work with
@KroganLab
and a large international group of scientists Spearhead by
@DoctorBou
@danishm20
and others
5
108
215
Now in peer-reviewed form - our study on network propagation of GWAS linked genes across >1000 human traits identifies aspects of cell biology that impact on multiple traits
@ibarrioh
et al. an
@OpenTargets
project
2
75
206
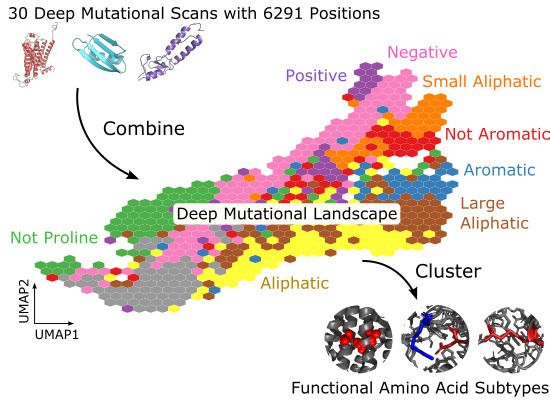
New lab preprint - Deep mutational scanning can tell us about the function of individual protein positions.
@Ally_Dunham
normalised DMS data for 6291 positions in 30 proteins to ask how many amino acid functions are there and how frequently are they used
3
69
163
New AI methods have led to the prediction of structures for hundreds of millions of proteins but these have to be explored. Together with
@thesteinegger
we clustered the 220 million structures in the
@emblebi
Alphafold DB to study their evolution and degree of novelty. See 🧵
We clustered the
#AlphaFold
structure database with our novel Foldseek algorithm. We identified 2.27M clusters and analyzed them by function, annotation, domains and evolution. Amazing collaboration with
@pedrobeltrao
lab. 1/n
📄
💾
11
273
842
1
30
148
Now in published form
@Cris_Vieitez
Bede Busby et al. on an experimental method to functionally annotate protein phosphorylation, using the power of yeast genetics (with
@TypasLab
,
@savitski_lab
and others at
@embl
)
5
45
144
EMBL will host a networking event for The Next Generation in Infection Biology. Apply if you are looking for PI positions in the area. >30 leading organisations will take part and registered their interest in you
(26-27 Jan 2022)
#EMBLInfectionBiology
5
88
144
Our institute
@emblebi
is looking to hire a group leader in computational biology (broad search). This comes with very generous core funding, access to HPC/GPU compute and the outstanding collaborative potential of EMBL, Sanger and Cambridge research
4
131
140
50 years ago the carnation revolution in Portugal led to the end of the longest running dictatorship in Europe. May this anniversary remind us that these freedoms we enjoy cannot be taken for granted
#25deabril50anos
1
24
133
Our (
@jurgjn
) review on recent progress in deep learning models for protein structure prediction and design is out in
@MolSystBiol
. This is meant as a primer for those outside wanting to get an update on what is going on with some thoughts on the future
0
42
130
Submitted the 10th preprint of our group to
@biorxivpreprint
today and it is fantastic that this is a common practice in a growing number of fields.
Years of research remain hidden from view without them. Don't let others live in your past. Preprint
3
32
128
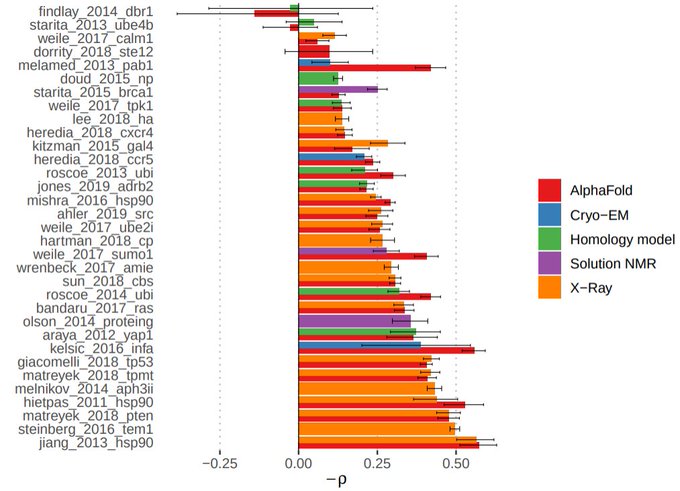
One final alphafold thing :) -
@Ally_Dunham
compared deep mutational scanning data with predicted changes in stability for mutations on the alphafold models (with FoldX). The correlations observed are typically as good or better than with experimentally derived structures
5
18
123
Our group is now formally affiliated with the Swiss Institute of Bioinformatics
@ISBSIB
. I look forward to contributing to the continued development of bioinformatics in Switzerland and to get to know our colleagues in joint meetings
4
6
121
The last PhD project from
@Ally_Dunham
was a collaboration with
@MoAlQuraishi
on a convolution neural network model for protein variant effect prediction. It achieves fast effect prediction without alignments.
3
19
107
I am very thankful for the Helmut Horten Foundation, that has supported my professorship appointment at
@ETH
. Through this I join
@theLoopZurich
, a medical centre for translational research and precision medicine
14
2
102
@EMBL
and
@DeepMind
have teamed up to provide 350,000 pre-computed structural predictions for 21 species using AlphaFold - fuly accessible via
EMBL and
@DeepMind
have partnered – a breakthrough for science.
Together, we're providing a treasure trove of protein structure predictions powered by
#AlphaFold
to herald a new era for
#AI
-enabled biology.
22
775
2K
1
44
99
our latest preprint:
@omarwagih
"Comprehensive variant effect predictions of single nucleotide variants in model organisms"
Describing the resource for H. sapiens, S. cerevisiae and E. coli
2
55
98
Now published in
@PLOSBiology
-
@dbradley534
's study of the evolution of kinase-substrate active site recognition. When during evolution did kinases learn to recognise different types of motifs ? (quick summary and future directions)
2
35
98
More cancer cell line screening data from
@CancerDepMap
at the
@broadinstitute
4,518 drugs tested across 578 (pooled) human cancer cell lines in a single dose and 1,448 positives re-screened in 8-point dose response. Many new anti-cancer activities.
2
48
95
A tiny contribution of our lab to this amazing effort to identify SARS CoV2 human protein interactions with a goal of finding new drugs. Achieved at an unprecedented speed for such a study.
Our paper is out on
@biorxivpreprint
describing our SARS CoV2-human protein-protein interaction map and drug predictions from the data. It was a honor to work with so many fantastic scientists around the world. -Nevan
1K
579
1K
54
25
90
Proteins that interact together are often relevant for the same traits and network analyses of GWAS genes can help identify trait relevant cell biology. Here
@ibarrioh
applied this to study 1002 traits defining a pleiotropy map of human cell biology
2
33
92
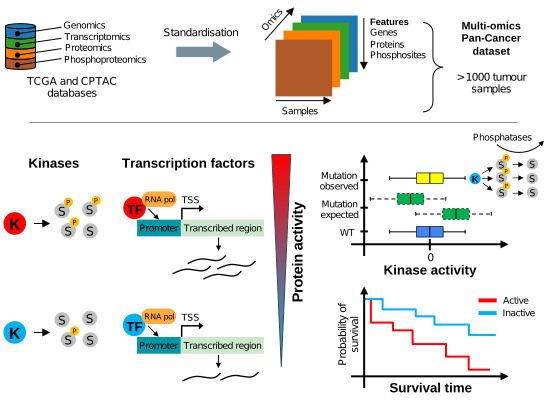
Cancer is more than its DNA. Now in published form, our study of predicted kinase/TF activities from multi-omics data, with a striking result being a lack of expected associations between mutations and activities
@abelfsousa
with
@DugourdAjf
@saezlab
@e_petsalaki
@danishm20
Estimation of activity changes for >500 kinases & transcription factors in >1,000 cancer samples & cell lines identifies drivers of signalling dysregulation & patient survival ➡️
@pedrobeltrao
@IMSB_ETH
@saezlab
@UniHeidelberg
@e_petsalaki
@emblebi
4
32
102
0
19
91
Congratulations to
@JulioSaezRod
@saezlab
on becoming the new head of research of
@emblebi
and congratulations to
@jomcentyre
for the promotion to Deputy Director of EMBL-EBI.
4
16
89
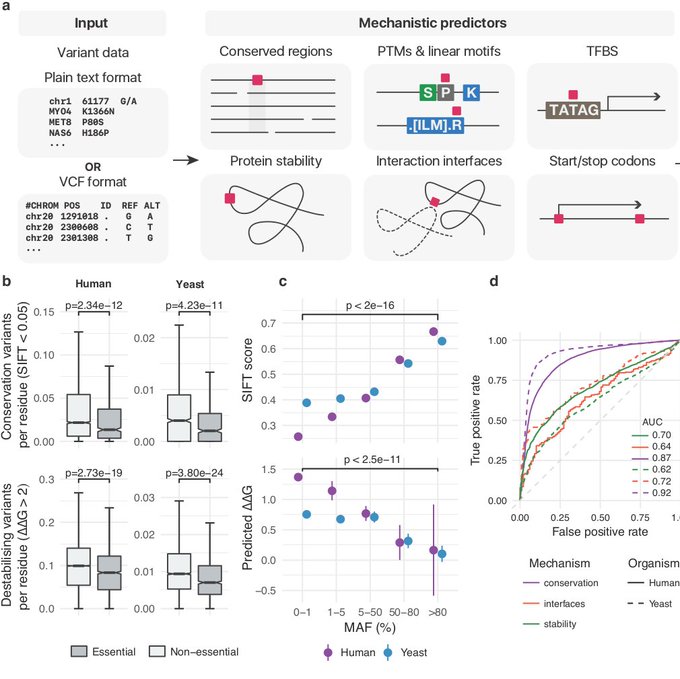
How often does a phenotype of a gene loss-of-function (LoF) depend on the genetic background ?
Now in published form - work from
@mgalactus
Bede Busby et al. studies the changes in gene deletion phenotypes across strains of S. cerevisiae
3
45
85
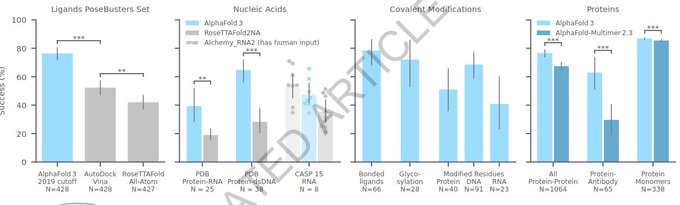
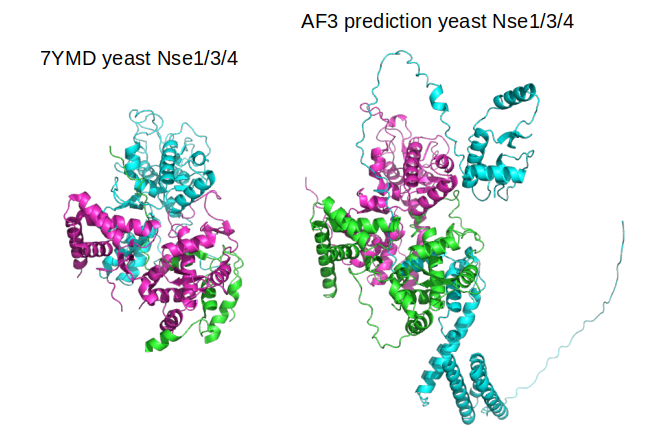
Companies like Deepmind do not have any requirements to make their research available. I signed the letter below because they chose to publish their work and different standards were applied to the Alphafold 3 publication as those applied to others.
We were incredibly disappointed with the lack of code or executable accompanying the publication of AF3 in
@Nature
. This contradicts scientific principles of the ability to evaluate, use, and build upon existing work.
4
126
410
2
16
86
AlphaFold predicted structures for the full protein universe (200 million). Quite a lot of redundancy in the set but so many discoveries can be powered by this. Novel enzymes, evolution of life, possibly protein dynamics from ensembles. The hart part is how to even analyse it all
Today in partnership with
@emblebi
, we’re releasing predicted structures for nearly all catalogued proteins known to science, which will expand the
#AlphaFold
database by over 200x - from nearly 1 million to 200+ million structures: 1/
107
2K
6K
3
26
82
I am pretty excited that our team with
@CorreiaMeloC
and
@KasperFugger
is among the selected to receive an HFSP research grant. We will be studying bioactive modified nucleotides and their modifying enzymes. It will also be great to be back at HFSP meetings.
🥳Congratulations to the 108 brilliant scientists awarded 2024
#HFSPResearchGrants
!🎉 Your pioneering work in
#lifesciences
is shaping the future of
#scientificinnovation
👏 Discover the visionary minds behind the breakthroughs:
#2024AwardeesBooklet
0
21
90
9
4
82
I had great day today at
@IGCiencia
where I will be periodically spending some time establishing collaborations along shared interests and helping to promote the application of computational approaches in life science research.
So happy to host the 1st
#GulbenkianSeniorFellow
@pedrobeltrao
. Pedro will spend regularly time at
@IGCiencia
to promote excellent science using computational biology approaches within the IGC and in Portugal aligned with IGC’s vision and mission!
2
2
59
2
4
81
In another effort by the QBI Coronavirus Research Group, we and many others have studied the commonalities of host targeted mechanisms of SARS-CoV-2, SARS-CoV-1 and MERS-CoV, identifying drug targets with potential pan-coronavirus activity
#QCRGglobal
1
36
80
We have a postdoc position open at
@IMSB_ETH
for someone with experience in phosphoproteomics in areas related with the function, dynamics, evolution and misregulation of PTMs (apply by March 20th, flexible starting date)
#TeamMassSpec
1
61
76
Very happy to join the Advisory editorial board of
@MolSystBiol
A warm welcome to our new Advisory Editorial Board members, we are looking forward to working with you! 🎉
And... a good occasion to thank our entire AEB for their support!
#SysBio
#SystemsBiology
2
7
31
1
0
75
Building phylogenetic trees guided by predicted protein structures, made possible by describing proteins with the Foldseek structural alphabet. I think this will also have an impact on ancestral sequence/structure reconstruction.
In a post
#AlphaFold
world, we can use protein structures in ways we never could before. Can we build phylogenies with them? Are they any good? Yes! Foldtree () surpasses traditional sequence-based methods, even for closely related proteins.👇
11
255
768
1
12
74
Thousands of phosphosites have been discovered with <5% having a known function. In our new preprint
@d0choa
uses a machine learning approach to address this gap
If you care what phosphosites may regulate your protein/process of interest read on
5
42
72
The
@emblebi
Alphafold database was just expanded to double the size with new predictions for most of the manually curated
@uniprot
entries. It will continue to expand over next year to cover millions of protein sequences.
🎉New
#AlphaFold
data! With
@DeepMind
, we’ve more than doubled the size of the database & added predictions for most of the manually-curated
@uniprot
entries in UniProtKB/SwissProt.
That's >400,000 new protein structure predictions for you to explore!
7
188
585
1
14
74
My group at
@IMSB_ETH
ETHZ is recruiting a lab technician/manager with a PhD in S. cerevisiae genetics / cell bio. ~60% time devoted to research on the functional relevance of PTMs and genetics of trait variation and ~40% on lab management
0
61
72
The last PhD project from
@Ally_Dunham
is now out in published form. A collaboration with
@MoAlQuraishi
where Ally developed a very fast protein missense variant effect predictor using a convolutional neural network model.
1
19
71
Large cancer studies have been sequencing based despite it being a signalling disease. Here
@abelfsousa
compiled matched genomic and (phospho)proteomic data for 1,110 tumours and estimated activity changes in ~500 kinases/TFs based on their targets.
2
21
70
I am passing all our future abstracts through this thing. It is brutal but surprisingly informative .
1
14
66
In February of last year Roman Cheplyaka (
@bioinfochat
) recorded a chat with me,
@janani_hex
and
@_amelie_rocks
on AlphaFold 2 but then his country was invaded. Somehow he managed to find time (!) to edit the first episode which you can find here
0
13
66
Just out in
@molcellprot
is
@abelfsousa
's PhD project studying why gene copy number changes are attenuated at the protein level and the implications this has on linking eQTLs to phenotypes. (w
@emanuelvgo
@BMirauta
@d0choa
@OliverStegle
)
2
24
64
New preprint
@Cris_Vieitez
Bede Busby et al. on our biggest effort to date to functionally annotate protein phosphorylation, using the awesome power of yeast genetics (with the Typas and
@savitski_lab
and others at
@embl
)
2
27
62
Now in published form -
@Ally_Dunham
combined experimental outcomes of mutations for 6291 positions in 30 proteins, showing these can group into different functional groups. Ally then studied these in the context of protein structures and evolution.
#DeepMutationalScanning
data (covering 6,291 positions in 30 proteins) are combined & the consequences of mutation at each position are used to define a mutational landscape & explore aa functions -->
@pedrobeltrao
@Ally_Dunham
@emblebi
#ProteinStructure
1
25
75
1
16
60
Out today, work by
@dbradley534
@Cris_Vieitez
and others, tries to identify which residues in protein kinases determine target preferences and how these rules work. Jointly with
@pedro_cutillas
lab th
https://www.cell.com/cell-reports/fulltext/S2211-1247(20)31591-6
2
20
59
Together with the lab of
@mjamorim1
at
@IGCiencia
we are hiring 2 postdoctoral researchers in Lisbon - bioinformatics (structural, networks, omics) to study viral proteins/networks and to help develop new therapeutics. Application instructions (PDF !)
0
26
58
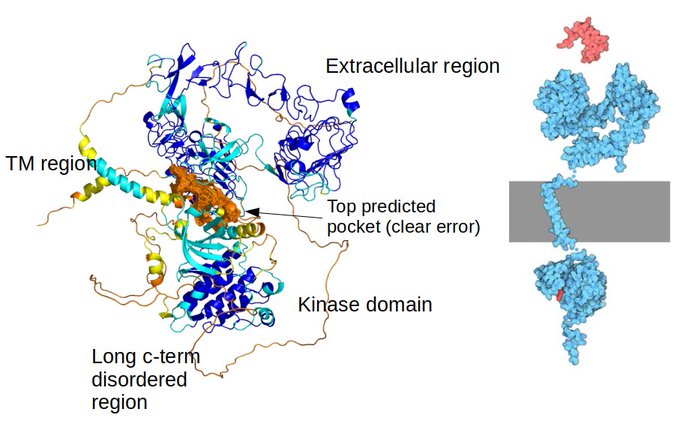
We are playing with the alphafold models like many others. Here are some generic issues using as example the EGFR based on work by
@jurgjn
that is evaluating the models for pocket detection and docking.
1
18
58
Now in press
@mgalactus
et al. Predicting E. coli strain & condition specific growth phenotypes from genomes (jointly with Typas lab)
1
38
54
Now in published form -
@danishm20
identified reproducible associations between gene copy number variation (CNVs) and phosphorylation changes across tumours. (jointly with
@MillerLab4
)
3
20
57
Registrations open:
@EMBO
Seefeld signalling meeting (11-16 Sep). Bringing together people with different mindsets (structures to organism, systems to engineering). It has a great list of invited speakers and many talks will be selected from abstracts
1
13
55
The increased focus in exome sequencing and rare coding variant with higher effect sizes is going to shift human genetics into protein and cell biology (vs. gene expression regulation). I find that exciting as it is easier to follow up and more likely to result in therapies
0
4
54
A proteomics approach to identify interactions regulated by phosphorylation. Part of the growing arsenal of tools to annotate the functional importance of protein phosphorylation
1
17
55
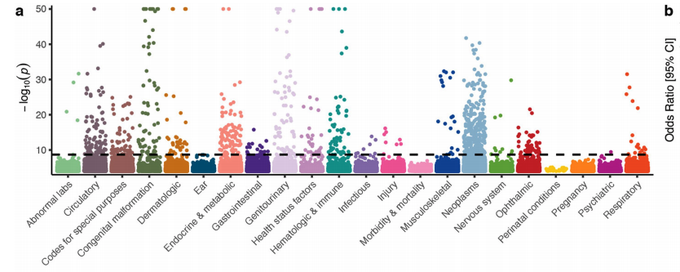
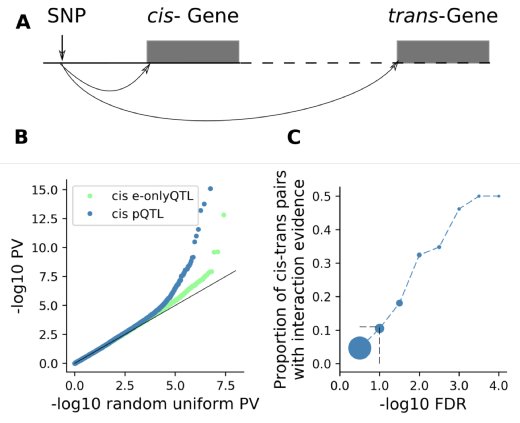
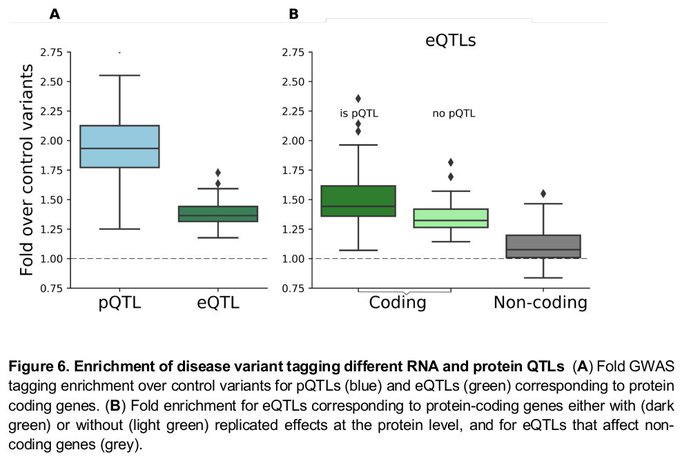
Proteomic study of 202 human iPSC lines with matched mRNA and genome information:
- eQTLs can result in trans protein QTLs via protein interactions
- pQTLs are more enriched in disease-linked GWAS variants than eQTLs
@OliverStegle
& Lamond labs
0
33
54
Finally, this is very much a
@Twitter
paper. It was born from the early access of results here and initially coordinate here before we moved to google docs and email.
0
4
54