Hung Nguyen
@tienhung_91
Followers
297
Following
611
Media
17
Statuses
90
Assistant Prof @UBChemistry | PD @UTChemistry | Computational modeling of RNA structure and folding, intrinsically disordered proteins and phase separation
Buffalo, NY
Joined October 2015
Looking to fill 1 or 2 postdocs who are interested in developing computational models to investigate structures & dynamics of biomolecules! Check lab website for what we're up to Job details here Please RT!.
0
9
13
RT @NaotoHori: With @ThirumalaiArmy and @tienhung_91, we wrote a review on 'Minimal Models for RNA Simulations', now published in @COSB_CRS….
0
6
0
Excited to share our lab's first preprint! We found that flexible RNAs have much broadened ion atmospheres than structured RNAs. This leads us to speculate the implications on molecular recognition and phase behavior. Please check full text for other interesting details.
We are happy to share our recent work entitled "RNA structural complexity dictates its ion atmosphere" on bioRxiv (doi: . Thanks to @tienhung_91.
1
0
0
lol, got me for a minute.
Since we're talking about funding of absurd research by NIH and other federal agencies, the funded scientists:. - watching flies fuck.- giving rats massages.- spending years digging into why jellyfish glow.- tracking penguin poop from space.- using horseshoe crab blood.
0
0
0
RT @sucheol_shin: RT plz! APS Focus Session "Genome Organization & Subnuclear Phenomena" is coming back at the 2025 Global Physics Summit!….
0
7
0
For those interested in grad school, @UBuffalo @UBChemistry has a virtual open house Nov 16 12:00 ET. Register below to learn more about our department! Link:
0
2
6
RT @HIRANMAY67: Happy to share our recent work on @JPhysChem Lett (. Thanks to @davethirum, @tienhung_91, @NaotoHor….
pubs.acs.org
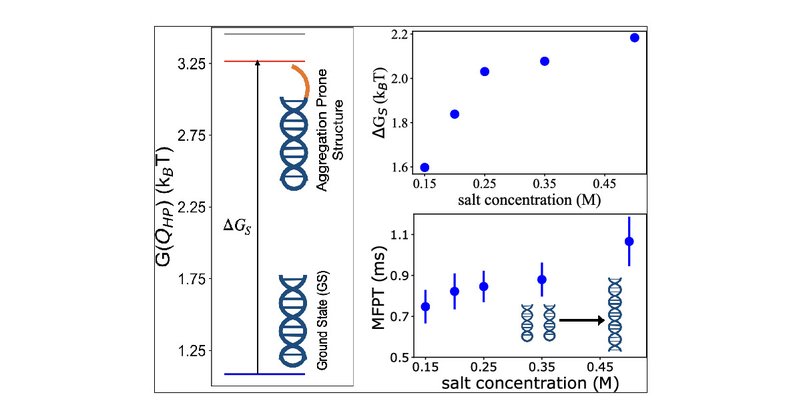
Repeat RNA sequences self-associate to form condensates. Simulations of a coarse-grained single-interaction site model for (CAG)n (n = 30 and 31) show that the salt-dependent free energy gap, ΔGS,...
0
3
0
Interested in applying to #Chemistry grad study? Learn about what @UBChemistry can offer in a virtual informational session. Ask us anything (including how to make such a cool poster)! Free to join and everyone is welcome! Pls RT. Register link
0
8
13
Our first ever (and certainly not the last) group lunch to welcome Peter @pz_chem_tweet joining the lab! Super excited to have my first grad student!! Looking forward to working with you Peter!
1
0
23
RT @SarbajitBanerj1: We are hiring! Check out three open positions @TAMUChemistry .Physical/Analytical Chemistry -….
0
28
0
Close one page, open another. Privileged to work with Dave and amazing colleagues @ThirumalaiArmy. Learnt a lot from you!!.
We had a farewell party for @tienhung_91 who starts his independent career at @UBChemistry. Best wishes for Hung and his future!
0
0
17
RT @MeyerLabDuke: Interested in using new technologies to study how RNA modifications regulate gene expression? Come join our team! The Mey….
themeyerlab.com
The laboratory of Kate Meyer in the Department of Biochemistry at the Duke University School of Medicine studies RNA biology. We seek to understand how RNA modifications control gene expression, with...
0
47
0
RT @RommieAmaro: 🚨Want to learn about the latest advances in Computational Structural Biology? . Join our #EMBOComp3D workshop 🤓. in Heidel….
0
59
0
RT @lab_COSMO: Everything ready for the @cecamEvents/@Psik_Network workshop in Berlin, organized by @CecClementi with a little help from @M….
0
18
0
RT @fcrameri: Scientific colour maps 8.0. #useBatlow.#ScientificVisualisation #visualisation #Science #ColourPalet….
0
456
0
Our recent work has been accepted and now appears on @PNAS! We establish a link between structures & thermodynamics of monomeric #RNAs and their propensity to undergo phase separation & aggregation.
We are happy to share our recent work on @PNASNews (. Thanks to @davethirum, @tienhung_91, @NaotoHori and @ThirumalaiArmy . We established a link between the folding landscapes of the monomers of CAG repeats RNA and their propensity to self-associate.
0
1
10
RT @BanerjeeLab_UB: We are hiring! Please RT. Dive into the fascinating world of #Biophysics as a Postdoc at Banerjee Lab, .
banerjeelab.org
0
87
0
6/n This work is spearheaded by the great twitter-less Mauro Mugnai, with contributions from @abhinawkumar21, @debayanbiophys, @ZenoResearch, @StachowiakLab, John Straub and of course @davethirum. Comments and feedback are welcome!.
0
0
1
5/n Want to try our model, check out the code here (. Install OpenMM, and provide the sequence, temperature, salt concentration and it should work out of the box. That's it!! Yes, seriously! Oh, and it will autopick GPU if available to speed up simulations.
github.com
Contribute to tienhungf91/SOP_IDP2 development by creating an account on GitHub.
1
0
1