gagneurlab
@gagneurlab
Followers
1K
Following
111
Media
67
Statuses
443
News from the Gagneurlab@TUM -- To understand the genetic basis of gene regulation and its implication in diseases.
TUM @ Munich, Bavaria
Joined February 2017
Starting in 15 min!.
Join us for our next Kipoi Seminar with with Dmitry Penzar, @dmitrypenzar @ .👉LegNet: parameter-efficient modeling of gene regulatory regions using modern convolutional neural network .📅Wed Dec 4, 5:30pm CET.🧬 🦋@kipoizoo.bsky.
0
0
0
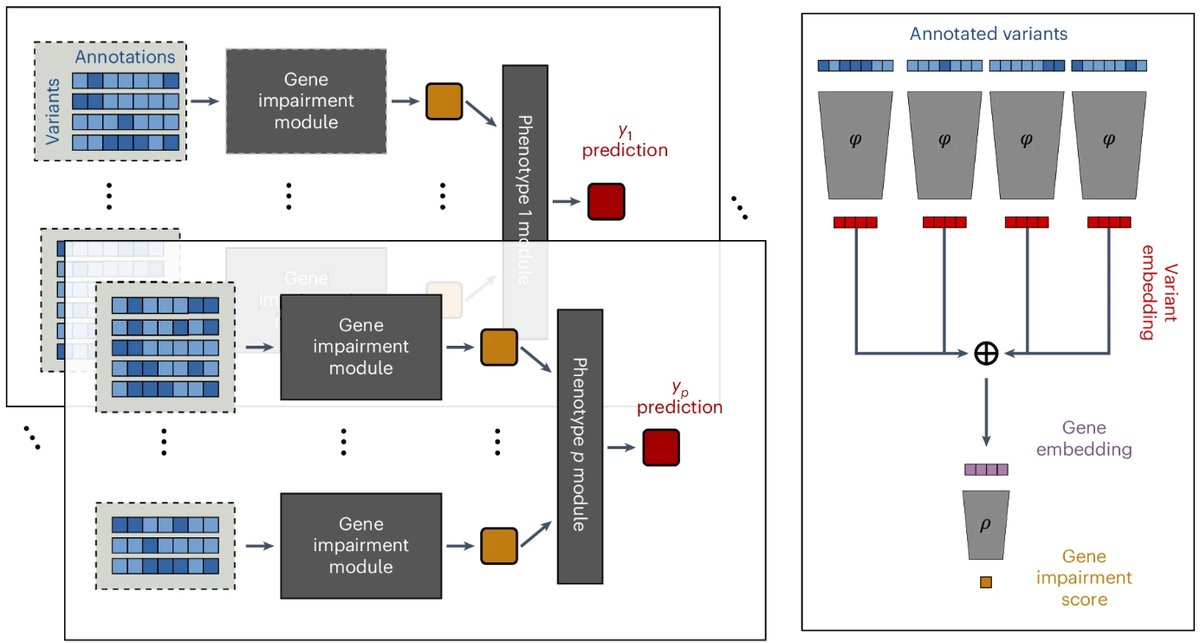
Tomorrow at #ASHG2024, Shubhankar @sg_londhe will present FuncRVP, our model improving rare variant testing by integrating gene impairment scores from DeepRVAT with functional gene information. Don’t miss it: we got fresh results since our July preprint!
Looking to enhance rare variant association testing? Catch my platform talk on FuncRVP - a Bayesian rare variant association framework leveraging functional gene embeddings. Nov 7, 11:30-11:45, Room 501. #ASHG24
0
2
3
Information leakage due to homology: A pervasive and non-trivial issue in bioinformatics. Excited to hear about @muntakim_rafi's insights and recommendations for sequence-based modeling on Wednesday at the Kipoi seminar.
Join us for the next Kipoi Seminar with with Abdul Muntakim Rafi (Rafi) @muntakim_rafi @CarldeBoerPhD @UBC! .👉Detecting and avoiding homology-based data leakage in genome-trained sequence models .📅Wed Nov 6, 5:30pm CET.🧬
0
5
12
Honoured to have written this review with Jernej Ule’s lab on the splicing code, spanning modeling, mechanisms and its implication for therapeutics designs. Wonderful teamwork.
From computational models of the splicing code to regulatory mechanisms and therapeutic implications #Review by @tiny_captain, @OscarWilkins16, @nils_f_wagner, Julien @gagneurlab & Jernej @ule_lab.@HelmholtzMunich @TU_Muenchen @TheCrick @KingsCollegeLon.
0
6
23
Avantika Lal @lal_avantika today at the Kipoi seminar - modeling single-cell gene expression in health and disease from sequence. Very much looking forward!.
Join us for the next Kipoi Seminar with Avantika Lal.@lal_avantika @genentech!.👉Decoding sequence determinants of gene expression in diverse cellular and disease states.📆Today Wed Oct 2, 5:30pm CET, not recorded! .🧬 Registration:
0
0
6
RT @OliverStegle: Really pleased to see DeepRVAT published. A new method for rare variant genetics by @Holtkamp_Eva & @brianfclarke in coll….
0
12
0
RT @OpenBoxSci: OBS #Genomics Seminar.📆 Thursday, 9/26, 11 am ET/5 pm CET . 🎙️ .Miquel Anglada Girotto.@m1quelag @CRGenomica .@lab_serrano….
0
4
0
DeepRVAT now out! High potential for rare disease res.: a) applies to very imbalanced case-control settings, b) our gene impairment score is trained w/o the pervasive disease-gene list bias: the strength of training on UKBB. Gr8t work @brianfclarke @Holtkamp_Eva w @OliverStegle.
🧵 1/9 Delighted to announce that #DeepRVAT is out today in Nature Genetics! Here's a recap of our tweetorial from the preprint, with updates for some exciting new results. It's been a joy to work with @Holtkamp_Eva, as well as @gagneurlab and @OliverStegle.
0
0
10
Taking sequence-based modeling to the next level: 500 kb DNA input, single-cell resolution, joint ATAC & RNA coverage tracks. Thumbs up to Johannes @thisisjohahi & Laura @lauradmartens. Wonderful collab with @JD_Buenrostro & @fabian_theis. Open source, of course. Enjoy!.
Excited to present “scooby”, which models multi-omic profiles (scRNA-seq coverage & scATAC-seq insertions) directly from 500 kb DNA sequence at single-cell resolution. This was a fantastic collaboration co-led with @thisisjohahi.
1
10
58
Proud to be part of the European Rare Diseases Research Alliance @ERDERA_org. Looking forward to fruitful collaboration across Europe to accelerate and increase the rate of RD diagnostics.
The European Rare Diseases Research Alliance @ERDERA_org kicks off with a bold vision to make Europe a global leader in rare diseases research and innovation. 🚀. Read press release🗞️. 🌍💙 #ERDERA #RareDiseases . @EUScienceInnov @HorizonEU @Inserm
0
0
10
RT @KipoiZoo: Join us for the next Kipoi Seminar ft. Max Horlbeck & Ruochi Zhang (@RuochiZhang) from @JD_Buenrostro's lab at @Harvard & @b….
0
2
0
New concept for variant effect prediction. 'How bad' is the new 'how rare'. With DNA LMs we can ask how important a variant is for predicting other bases. This influence score correlates better with functional assays. This was not possible with alignment-based conservation.
We show that nucleotide dependencies indicate deleteriousness of human genetic variants more effectively than sequence alignment and DNA LM reconstruction. Example: promoter mutagenesis screen from Kircher et al.
0
2
20
Thrilled to share our work on nucleotide dependencies. We can see the base relationships DNA language models capture. TF motifs and their interactions, splicing, RNA structures. the breadth of biology unraveled is awesome. And it matters for variant effect prediction! Enjoy.
Have you ever wondered what the genome looks like through the eyes of a DNA language model? In our newest preprint we use DNA LMs to study nucleotide dependencies in the genome, revealing functional elements, characterizing variants and evaluating DNA LMs
0
3
28
After DeepRVAT, FuncRVP is our next rare variant association testing tool! Rather than testing one gene at a time, we leverage information across genes using foundation models of gene function. All open source. Shubhankar Londhe @sg_londhe led this cool work - more in his thread.
1/n Do you want to push rare variant association testing to the next level? Check out our preprint of FuncRVP, a Bayesian rare variant association framework leveraging functional gene embeddings.
0
2
29
RT @anshulkundaje: A nice example of how training set design can have huge impact on generalizability. This continues to be an underappreci….
0
4
0
RT @lisamar2017: if you are passionate about the therapeutics of the future and want to be part of the 'RNA revolution' apply for this proj….
0
12
0
RT @AnneOtation: We explored why pathogenic variants are present in the gnomAD database for early-onset, severe, dominant conditions. Expla….
0
38
0
RT @lisamar2017: Join us for an exciting afternoon of RNA talks (including @ggiuliacantini from my lab) on the 10th of July at MPI for Bio….
0
2
0
Sequence design at the upcoming Kipoi webinar on Wed :-) Looking forward to @SagerGosai's talk!.
Join us for our next Kipoi Seminar with Sagar Gosai.@SagerGosai @r_tewhey @jacksonlab! .👉Machine-guided design of synthetic cell type-specific cis-regulatory elements.📆Wed July 3, 5:30pm CET .🧬
0
1
3