Explore tweets tagged as #Squidpy
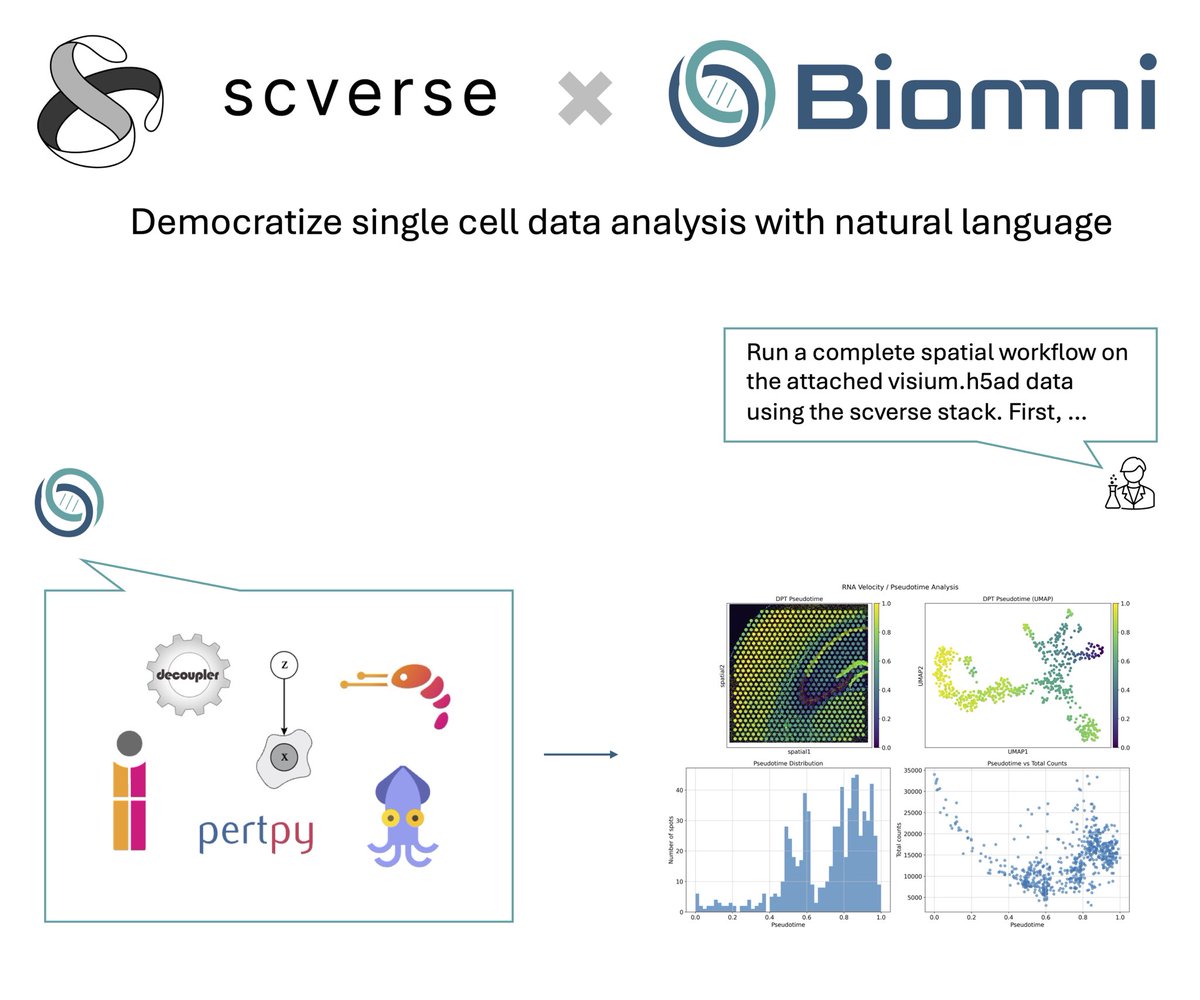
Thrilled to share @scverse_team x @ProjectBiomni! .Biomni agent now supports 10 scverse core packages including scanpy, squidpy, scirpy, pertpy, etc! .You can now use natural language to unlock complex single cell, spatial, and perturbation data analysis to generate novel
2
36
167
🤝Excited to announce @scverse_team X @ProjectBiomni!. Biomni now supports 10 core scverse packages—including scanpy, squidpy, scirpy, and pertpy—enabling researchers to perform single-cell, spatial, and perturbation analyses using natural language.
6
22
119
#MERSCOPE experiments create a vast amount of #SpatialData! Stop by our #SciPy2023 poster tonight at 6PM CDT to learn about our #CORMERANTlibrary that helps researchers navigate complex #MERFISH datasets. Learn more: #SCANPY #Squidpy @scverse_team
0
16
55
Why introduce a new platform for #scomics analysis when there's already Scanpy (+ its relative Squidpy), Seurat, Giotto, semla etc.?.First, @LambdaMoses realized there's a need for facilitating the use of methods from geospatial statistics that these platforms don't support. 2/
2
2
13
Ngl! messing with squidpy from @fabian_theis's lab during a lunch break has been fun . now back to pytorch!!! have a good Monday everyone!!!. #SpatialTranscriptomics #DataScience
2
0
5
The #OpenSource #spatialomics Github repos with the most activity were Giotto@suite_dev, spatialLIBD@master, squidpy@main, and Voyager@devel. - Giotto releases v3.2.1 on 'suite_dev' branch.- spatialLIBD releases v1.11.7 on 'master' branch. 🧵4/4
0
1
2
Now @HannahSpitzer1 is demonstrating the extensive capabilities of their #Python framework #Squidpy for single cell analysis. #omics
0
0
5
10/12 We released CellCharter as a Python library compatible with the @scverse_team ecosystem and thus, libraries such as anndata, scanpy, squidpy, and scvi-tools. We are planning to implement a lot of new features in the next period.
2
1
5
We’ve designed it to be fast, flexible and easy to use:.- Works seamlessly with Scanpy, Squidpy and AnnData (@scverse_team compatible).- Plug-and-play any ST dataset.- Outputs spatially informed scores.- Geostatistics-inspired metrics to evaluate gene sets.
1
1
3
. calculates gene set activity scores similarly to AUCell by @steinaerts , and obtains a Moran’s I score for each gene set using squidpy. For example, these two gene sets showed high Moran's I values, and they clearly mark biological structures. 7/21
1
0
1
6/9 - Display network diagrams to explore spatial neighbors graph, e.g. from #squidpy (test it live on :
1
1
4
4/9 The Python library bridges existing solutions for multimodal analysis including @scverse_team Squidpy, Scanpy and scvi-tools. We welcome new collaborations and we aim at tailoring the infrastructure to the new needs and innovations of the community.
1
3
12