Simona Cristea
@simocristea
Followers
9K
Following
6K
Media
587
Statuses
5K
cancer genomics AI scientist; head of Data Science & AI and group leader @DanaFarber_Hale; research scientist @Harvard; phd @eth.🇷🇴🇸🇪🇨🇭🇺🇸
Boston 🇺🇸 & Zurich🇨🇭
Joined January 2016
scRNAseq cell type annotation is notoriously messy. Despite so many algorithms, most researchers still rely on manual annotations using marker genes In a new preprint accepted at ICML GenAI Bio Workshop, we ask if reasoning LLMs (DeepSeek-R1) can help with cell type annotation🧵
6
42
200
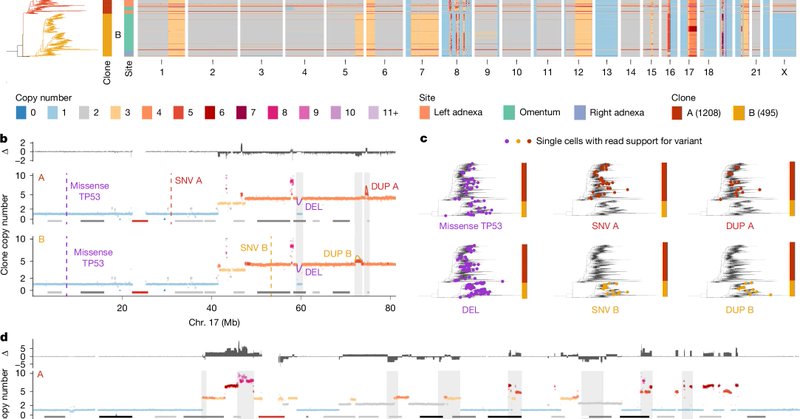
powerful ovarian cancer study @Nature “drug-resistant states in high grade serous ovarian cancer pre-exist at diagnosis, leading to positive selection and reduced clonal complexity at relapse. We suggest [..] evolution-informed adaptive treatment regimens to ablate resistance”
1
1
10
Hey @elonmusk! You were right again! Preschool Propaganda: Study Finds 41% of Netflix Kids Shows Include LGBT Themes @CWforA@YWforA https://t.co/OWkvwOFh5J
dailywire.com
It’s not in your imagination: children’s television has been overrun with LGBT themes, and Netflix is leading the charge, waving the rainbow flag right into your living room.A new report from...
21
45
316
great work about interpretability in PLMs using sparse autoencoders, similar to many of Anthropic’s investigations
How do protein language models (PLM) think about proteins?🧬 We answer this w/ #InterPLM, just published in @naturemethods! Using sparse autoencoders + LLM agent, we identify 1000s of interpretable concepts learned by PLMs, pointing to new biology 🧵
0
0
5
AIxBio’s best kept secret: computational biology = virtual cells
2
1
11
weird moment when a company works on a novel foundation model for one year and then they show…. a UMAP
0
0
3
🧬🔥
Ctrl-DNA, our constrained RL + Genomic Language Model system for cell-type–specific regulatory DNA design, was accepted as a @NeurIPSConf 2025 Spotlight (top 3.2%) 🧬✨ Paper: https://t.co/anDOO3xg9m Code: https://t.co/CQ9FPisCEk TL;DR We fine-tune DNA GLMs with a constraint RL
0
1
1
it’s crazy that so few people are able to clearly articulate what they need to get done so as to be able to prompt an LLM correctly
0
0
5
Google presents an AI system to write expert-level scientific software. Using LLMs + tree search, it invented novel methods in bioinformatics, epidemiology, geospatial analysis & more, often surpassing human SOTA. (1/4)
63
541
3K
“cancer-inspired computing as a paradigm drawing from the adaptive, resilient, and evolutionary strategies of cancer, for designing computational systems capable of thriving in dynamic, adversarial or resource-constrained environments”
1
0
4
just realized that most deep domain experts are very similar to reasoning LLMs: very knowledgable, most of the times right in their judgement, sometimes wrong, but unaware/unwilling to admit they are wrong 🤯
1
2
13
and they don’t like eachother
The AI field is now split into (A) a "traditional" ml/dl domain, and (B) a "psycho-AI" domain where innovation requires an understanding of / intuition about the cognitive capabilities of pretrained models and how to prompt / fine-tune them. These two fields are IMO separated.
0
0
3
RL scaling is meant to hit a wall in real-world tasks because such tasks are designed to be done by many people together & include lots of redundancies & inefficiencies. anybody who worked among many people knows that it’s seldom the “best” (by a clear metric) solution that wins.
0
0
4
extremely excited to share my lab's research of AI for cancer genomics at the neurIPS 2025 workshop on multi-modal foundation models in san diego
We are excited to organize NeurIPS 2025 2nd Workshop on Multi-modal Foundation Models and Large Language Models for Life Sciences. The workshop features a stellar lineup of invited speakers, including Ziv Bar-Joseph, Charlotte Bunne @_bunnech , Simona Cristea @simocristea ,
1
1
28
This R packages can transform any plot using color pallets from painting at the Museum of Modern Art in New York https://t.co/rUEaPru7N3
3
47
211