Kexin Huang
@KexinHuang5
Followers
1,823
Following
562
Media
50
Statuses
367
PhD Student @Stanford CS with @jure ; Machine Learning + Biomedicine
Joined August 2018
Don't wanna be here?
Send us removal request.
Explore trending content on Musk Viewer
オーロラ
• 492250 Tweets
Joost
• 334472 Tweets
#الهلال_الحزم
• 85607 Tweets
Fulham
• 81859 Tweets
#SixTONESANN
• 78237 Tweets
Man City
• 62845 Tweets
自分これ
• 61859 Tweets
Gvardiol
• 54232 Tweets
Ali Koç
• 47760 Tweets
Dremo
• 41197 Tweets
太陽フレアのせい
• 39467 Tweets
Burnley
• 33746 Tweets
SEE YOU NEXT MONTH JIN
• 31685 Tweets
دوري روشن
• 30597 Tweets
الدوري الاقوي
• 28769 Tweets
Granada
• 25210 Tweets
روشن السعودي
• 24591 Tweets
Luton
• 22315 Tweets
Haley
• 22196 Tweets
やまとなでしこ
• 14763 Tweets
الدوري التاريخي
• 14541 Tweets
#الهلال_بطل_الدوري_التاريخي
• 11927 Tweets
Last Seen Profiles
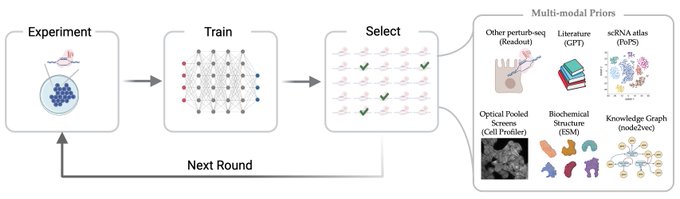
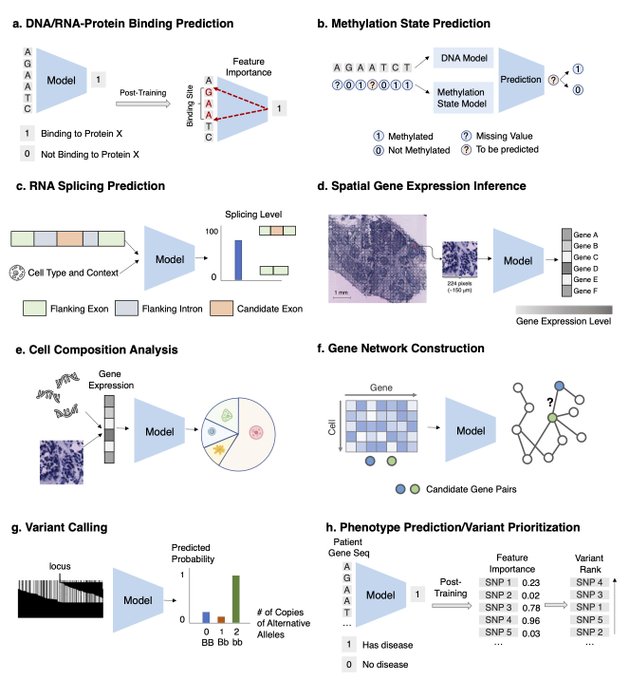
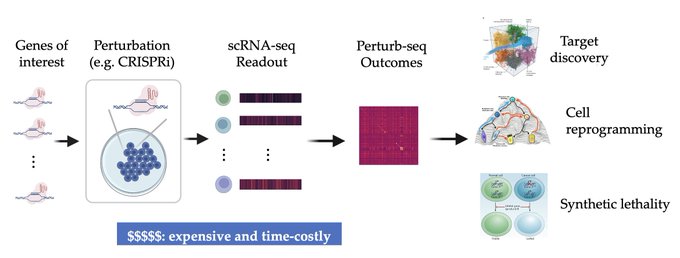
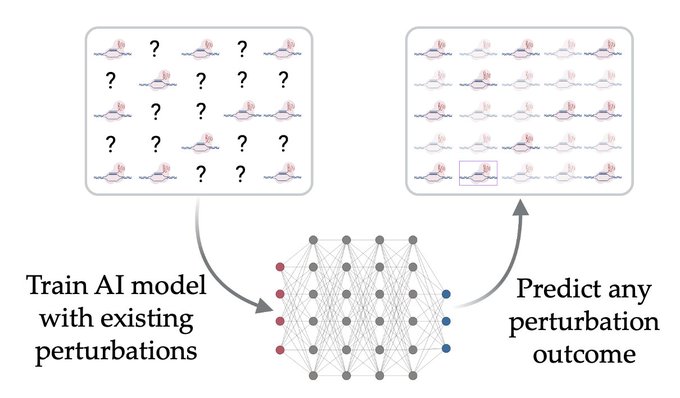
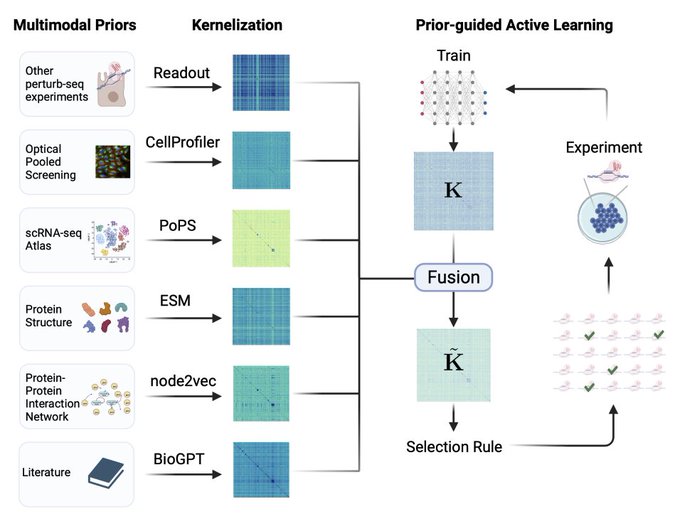
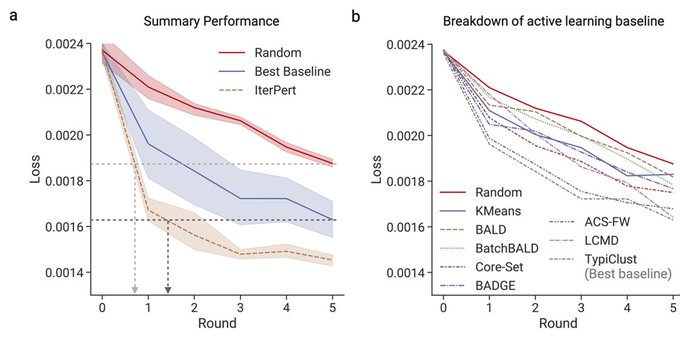
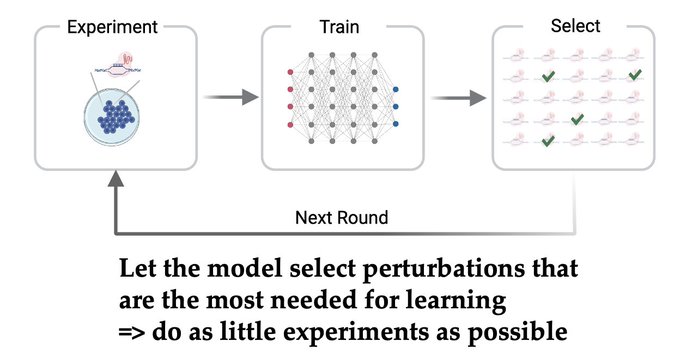
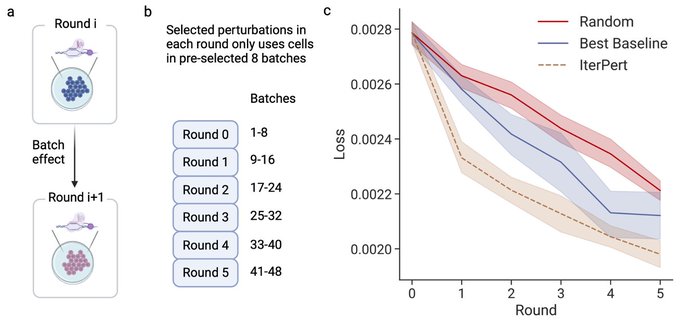
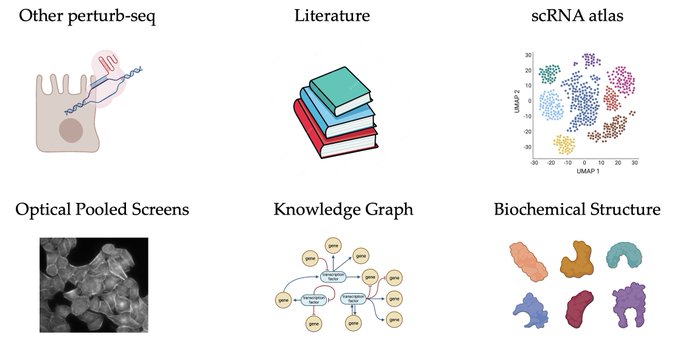
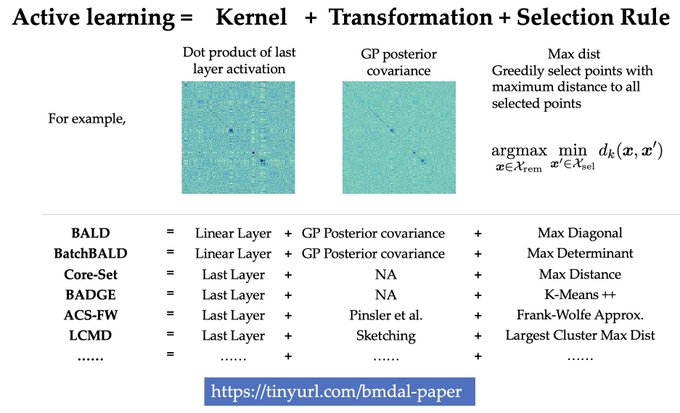
1/🧵Introducing Perturb-seq-in-the-loop: a sequential experimental design strategy for perturbation screens guided by multimodal priors, with 3X speedup over state-of-the-art active learning methods!
With amazing
@_romain_lopez_
@jchuetter
Taka Kudo
@antonio_science
Aviv Regev
7
113
473
An update ➡️ I will be attending
@Stanford
CS PhD program in the fall!! Super excited and grateful🙏🙏 Look forward to continuing research on machine learning + biomedicine!!
15
12
304
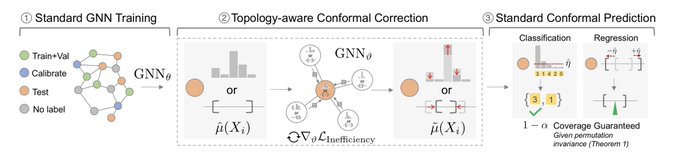
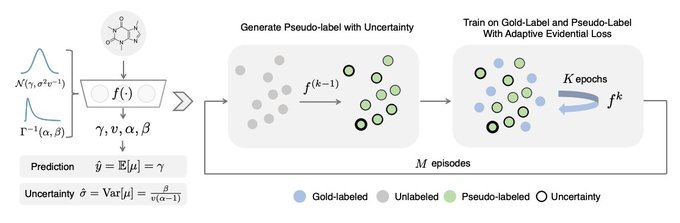
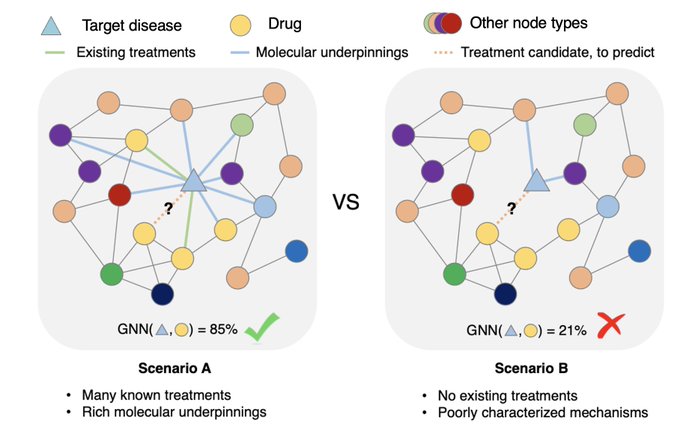
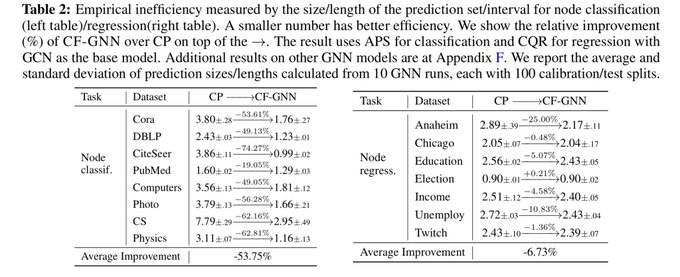
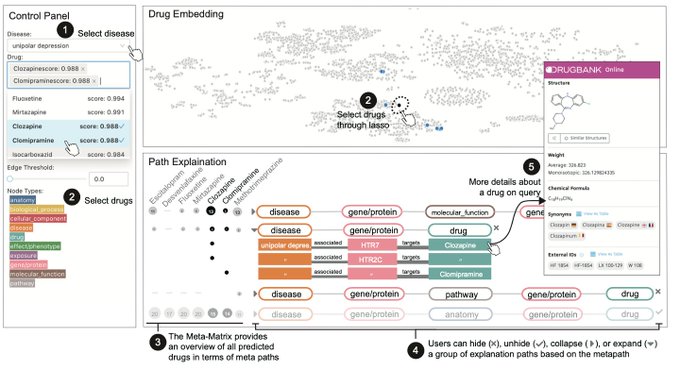
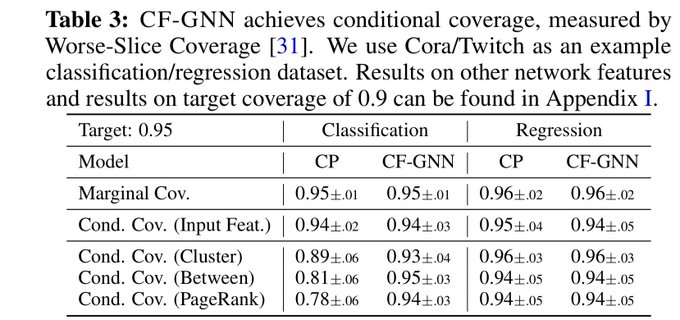
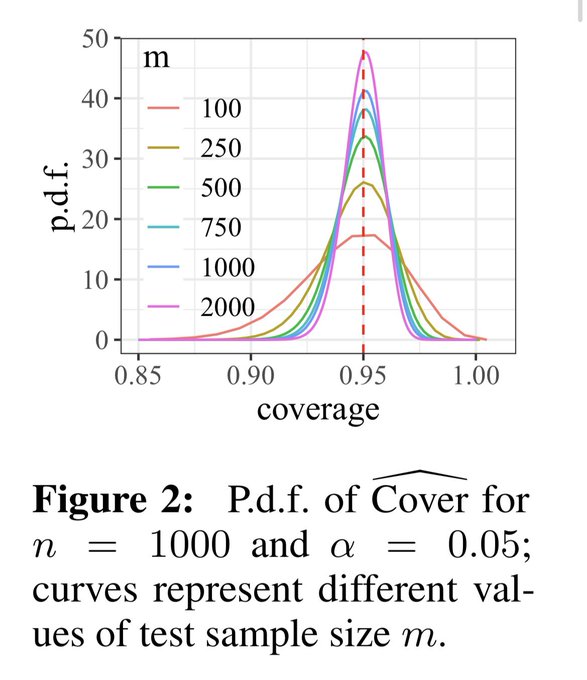
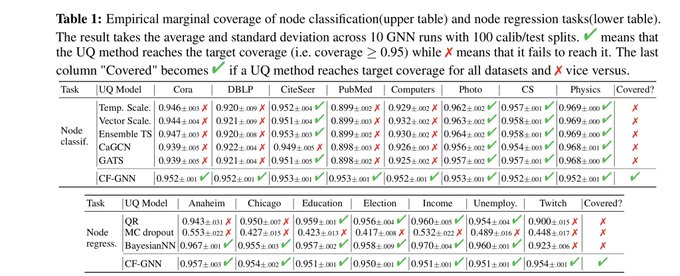
📣Excited to share our work on conformalized GNN with
@YingJin531
, Emmanuel Candes and
@jure
!
Given an entity in the graph, it produces a prediction set/interval that provably contains the true label with pre-defined coverage probability (e.g. 90%):
3
57
222
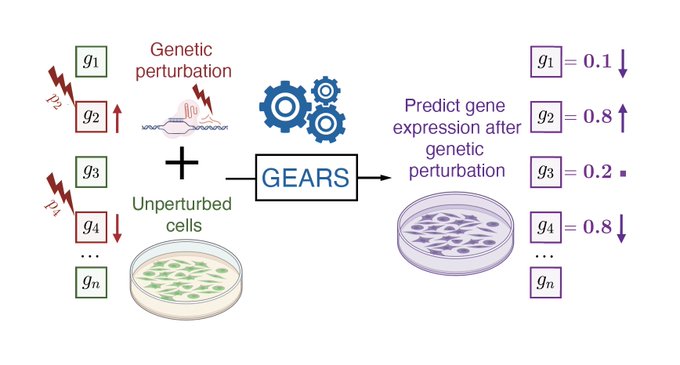
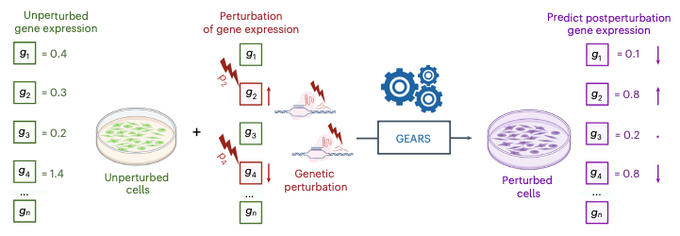
Excited to share our work on graph ML to model multiple and never-before-seen genetic perturbations! Tweetorial⬇️
0
20
167
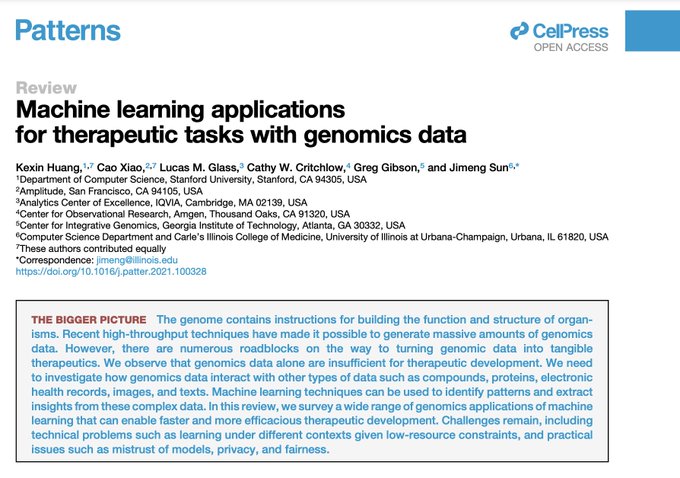
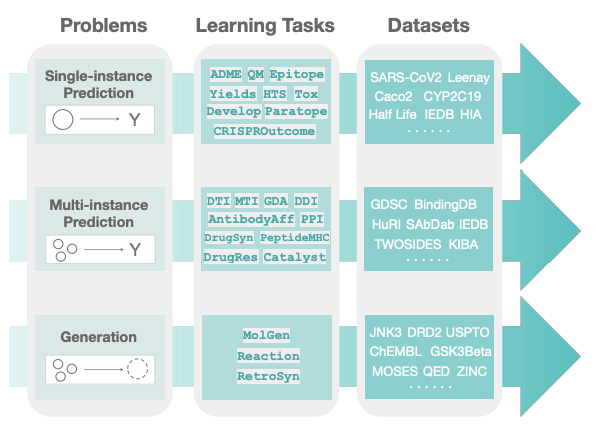
Excited to share a survey on ML for therapeutics tasks centering around genomics data
@Patterns_CP
!
We review 22 tasks across Tx pipelines that study the interplay of DNA seq, omics, compounds, proteins, texts, networks, and spatial data
4
30
151
Conformalized GNN to produce reliable graph predictions with statistical guarantees is now accepted by
#NeurIPS2023
as a spotlight!
Code is now available at
📣Excited to share our work on conformalized GNN with
@YingJin531
, Emmanuel Candes and
@jure
!
Given an entity in the graph, it produces a prediction set/interval that provably contains the true label with pre-defined coverage probability (e.g. 90%):
3
57
222
1
11
100
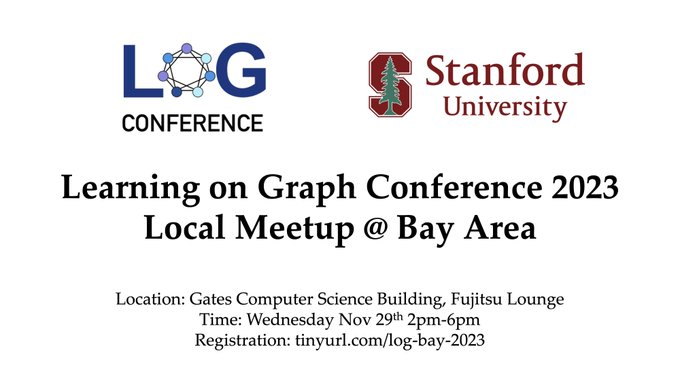
We are hosting a
@LogConference
Bay Area local meetup on Nov 29th! Come to Stanford to learn from and connect with the local graph learning community! Register here:
with
@ShirleyYXWu
@MinkaiX
@jure
0
24
93
Super excited to share our work on predicting unseen & combinatorial perturb-seq outcome using prior knowledge GNN
@NatureBiotech
lots of new results in the updated paper - check them out!
4
12
88
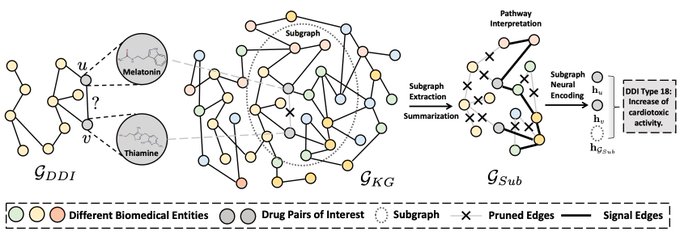
Happy to share SumGNN in Bioinformatics!
We inject biomedical knowledge from Hetionet KG and provide a scheme to generate potential mechanism pathways! Great results on DDI!
Paper:
GitHub:
Great work led by
@yueyu30308379
0
12
82
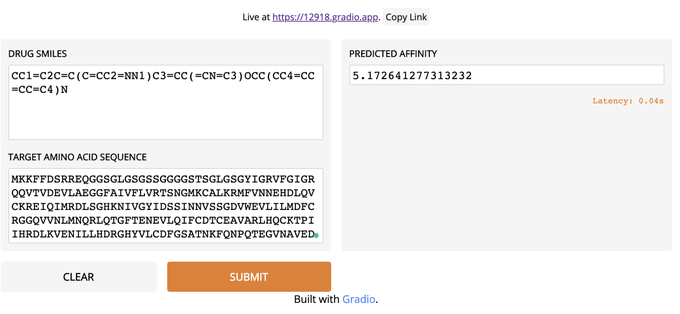
MolDesigner is in
#NeurIPS2020
Demo!
-Interactive molecule design with DL, powered by DeepPurpose and
@GradioML
!
-Predict binding affinity and 17 ADMET properties from 50+ DL models!
-Less than 1 sec latency!
Video:
Paper:
5
19
80
Thanks Ahmed for having me and really enjoyed the discussion! Here is also the link for the slide:
In our 3rd Conformal Prediction seminar,
@KexinHuang5
discusses his work w/
@YingJin531
, Emmanuel Candès, and
@jure
, exploring the application of conformal prediction to graph neural networks!
Watch the talk at:
0
9
64
0
5
69
We are hosting a
@LogConference
bay area local meetup! Come to Stanford to learn about exciting new research in LOG and hang out with graph ML folks! Register here:
with
@jure
@MinkaiX
@kaidicao
Lata Nair
0
11
69
I will be at NeurIPS presenting ⬇️ on Thursday poster session and also co-organizing
@AI_for_Science
workshop on Saturday! PM me if you would like to chat about anything related to AI + biological discovery!
📣Excited to share our work on conformalized GNN with
@YingJin531
, Emmanuel Candes and
@jure
!
Given an entity in the graph, it produces a prediction set/interval that provably contains the true label with pre-defined coverage probability (e.g. 90%):
3
57
222
1
2
59
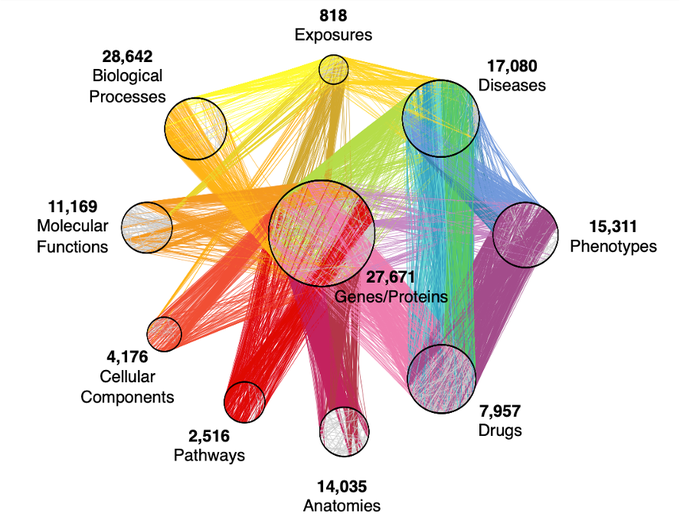
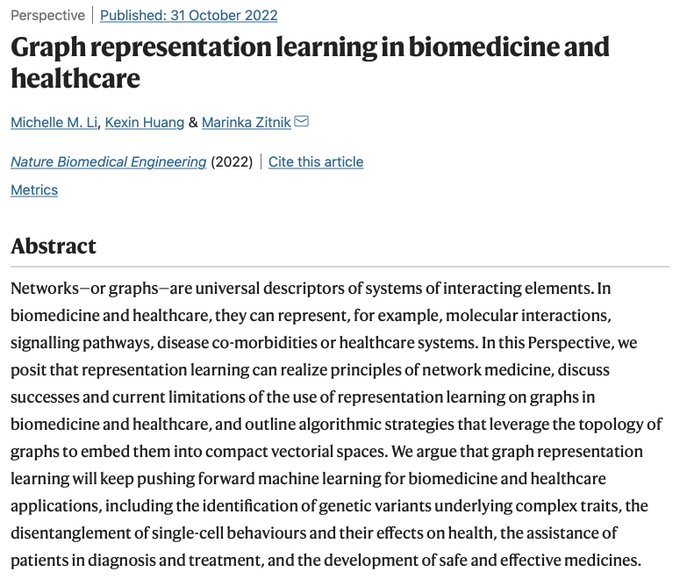
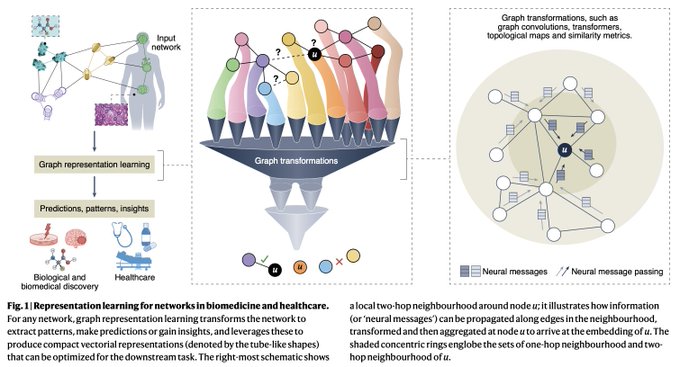
Very happy to share our perspective on the exciting intersection of graph ML + biomed & healthcare
@natBME
!
🙌We are beyond excited to share our Perspective on graph representation learning for biomedicine and healthcare!🥳
@natBME
@KexinHuang5
@marinkazitnik
@HarvardDBMI
(1/4)
6
37
214
0
7
51
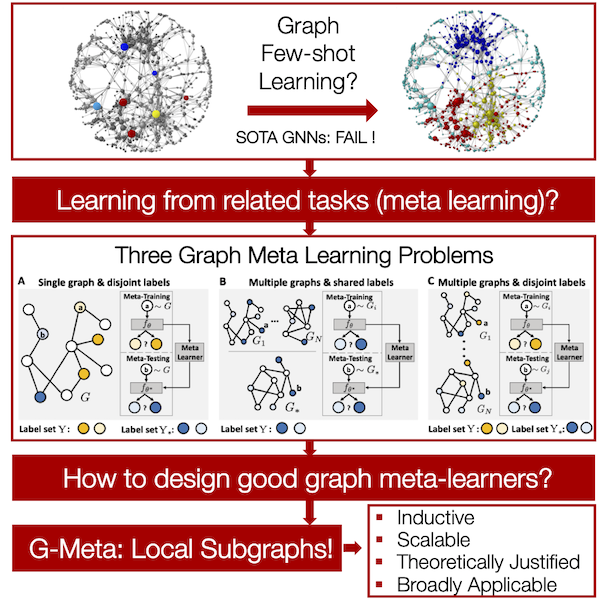
How to do node/link prediction when only a handful of labels are available? SOTA GNNs would fail!
Come to poster B1-C1 on Thursday 9pm PT
#NeurIPS2020
! ()
Project website: with
@marinkazitnik
!
0
15
46
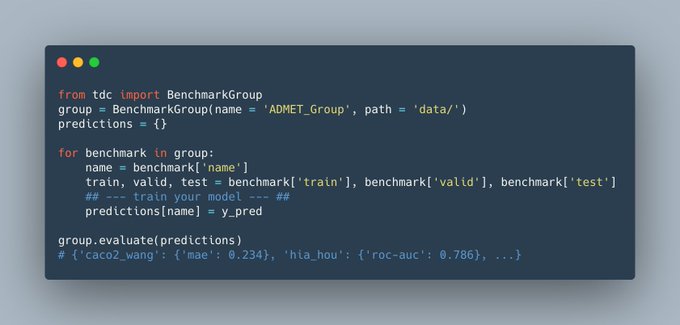
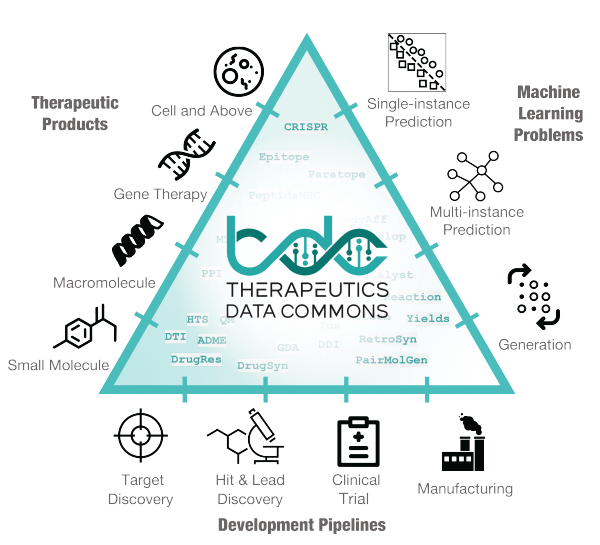
Very excited to share Therapeutics Data Commons, an ML data hub for therapeutics!
3 lines of code (!) to access
- 62 ML-ready datasets from 22 Tx tasks
- 10 Biologics Datasets
- 24 ADMET Datasets
- 20 Mol Oracles
Talks tmr 1:45pm EST at
#futuretx20
!
1
14
42
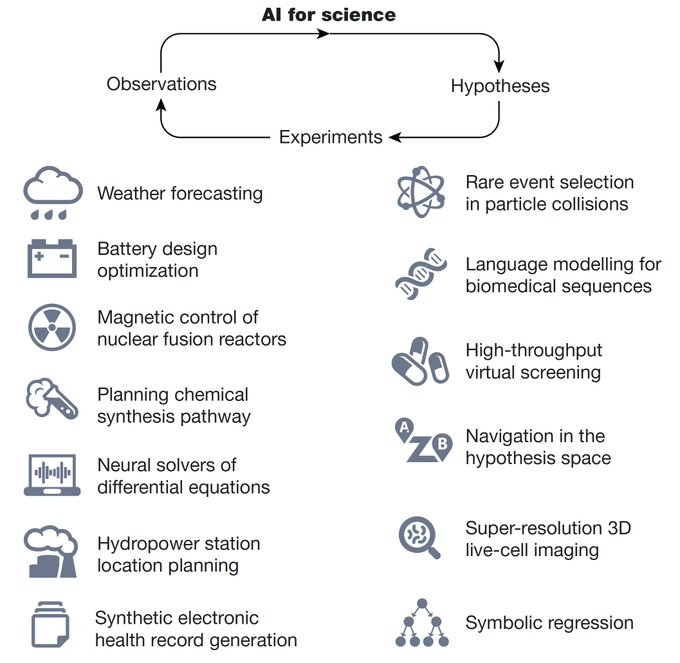
Excited to share our perspective on AI + Science! We review how AI accelerates the scientific inquiry loop (hypothesis -> experiments -> observations)
@AI_for_Science
How can
#AI
transform science?
Let us count the ways
A brilliant review
@Nature
@marinkazitnik
@TianfanFu
@YuanqiD
and colleagues
@AI_for_Science
#ScienceTwitter
12
104
330
0
5
42
Many international students don’t have access to resources on how to do proper research.
To fill the gap, with
@jimeng
and
@caoxiao_danica
, we initiate “Sunstella Mentorship Fellow” for PhD/PostDoc/… who are excited to help international junior students over summer 2022⬇️
2
8
39
Awesome blog about exciting graph ML works in the past year! Also checkout my reviews on the graph learning for system biology and robustness sections!
📣Two new blog posts - a comprehensive review of Graph and Geometric ML in 2023 with predictions for 2024.
Together with
@mmbronstein
, we asked 30 academic and industrial experts about the most important things happened in their areas and open challenges to be solved.
🧵 1/n
3
129
449
0
4
32
Excited to share our annual review on 🔥 research in AI for science from 2023!
1
1
32
Super excited to share TDC paper
@nchembio
- see tweetorial ⬇️
Excited to share our new paper in Nature Chemical Biology
@nchembio
AI is poised to transform
#therapeutic
#science
The Commons is an initiative to access and evaluate
#AI
capability across therapeutic modalities and stages of discovery 1/4
1
19
72
1
5
31
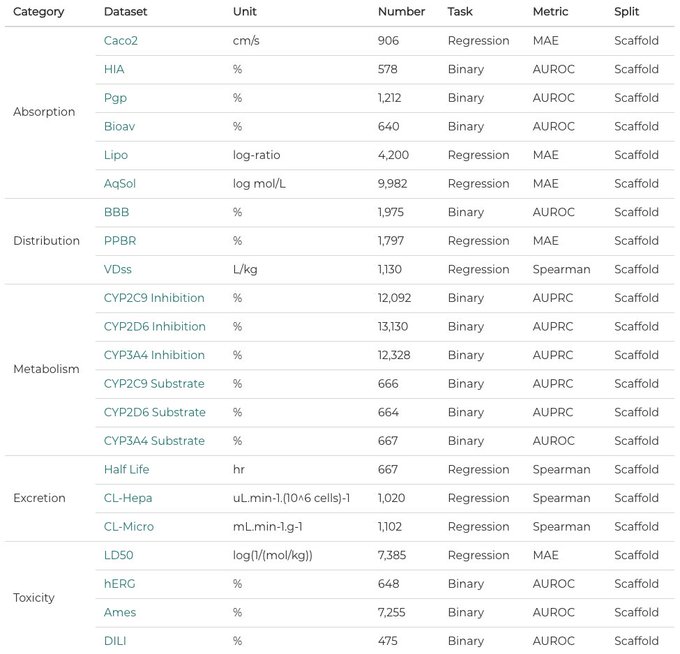
Happy to share the first leaderboard of TDC: 22 new & important ADMET property prediction datasets for molecular ML model! Submit your model to TDC!
More info:
GitHub:
@marinkazitnik
@TianfanFu
@WenhaoGao1
@yzhao062
@yusufroohani
0
9
28
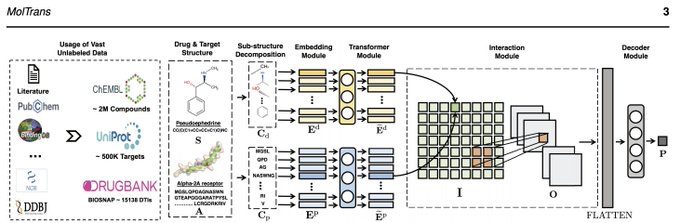
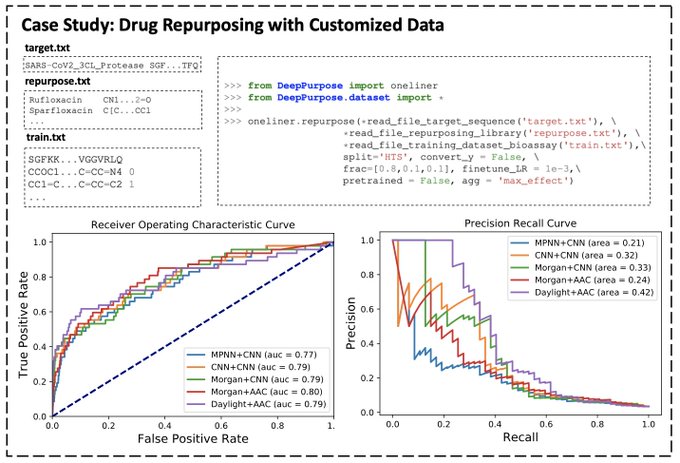
Checkout MolTrans, a new deep learning model for Drug-Target Interaction Prediction! Now published in Bioinformatics!
Code:
Paper:
Check out our paper of a new deep learning method for drug target interaction (DTI) prediction
@AiLucasg
@caoxiao_danica
@KexinHuang5
1
3
17
3
4
24
Excited to co-organize this workshop⬇️
Introducing AI for Science — a
@NeurIPSConf
2021 Workshop! Our workshop focuses on bridging the gaps between machine learning and science. We have a stellar lineup of speakers! Submit your work and sign up for mentorship program now:
2
52
189
0
1
24
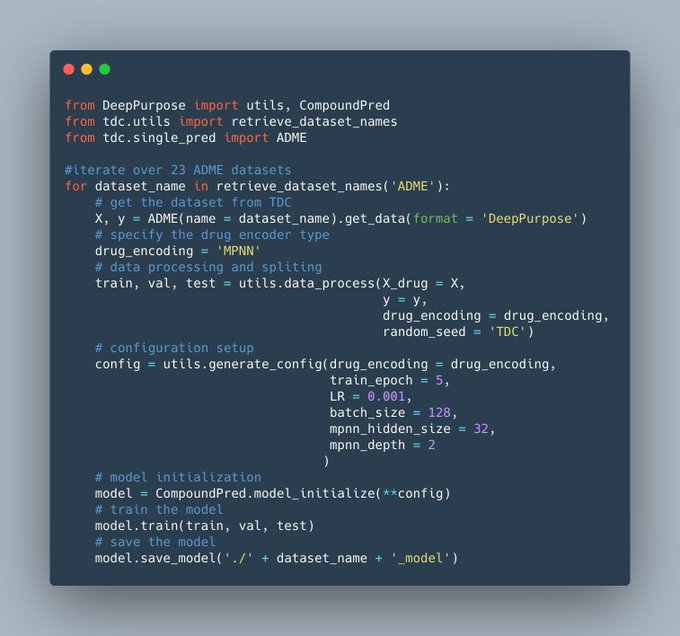
Just create a web UI for Drug-Target Interaction Prediction with less than 10 lines using DeepPurpose and
@GradioML
👇 Very impressed with Gradio's simplicity!
2
10
21
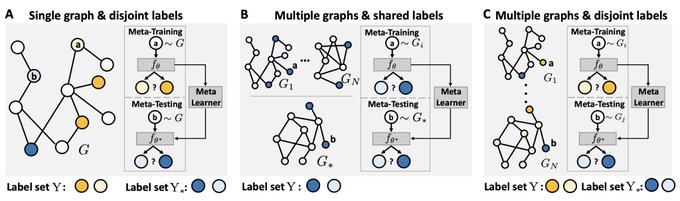
Excited to share G-Meta with
@marinkazitnik
:
- Local subgraph enables knowledge transfers in various graph meta-learning problems, with theoretical motivation!
- Promising result on 7 datasets, where two are new & large (1,840 graphs)!
- G-Meta scales!
-
0
6
21
Our new preprint on a comprehensive survey for graph learning + biomedicine! Lots of exciting opportunities: Link ⬇️
I am excited to finally announce our survey of representation learning for networks in bio. and med. w/
@KexinHuang5
&
@marinkazitnik
! We synthesize a spectrum of rep. learning approaches & share 4 unique prospective studies to demonstrate their potential!
1
8
38
0
1
21
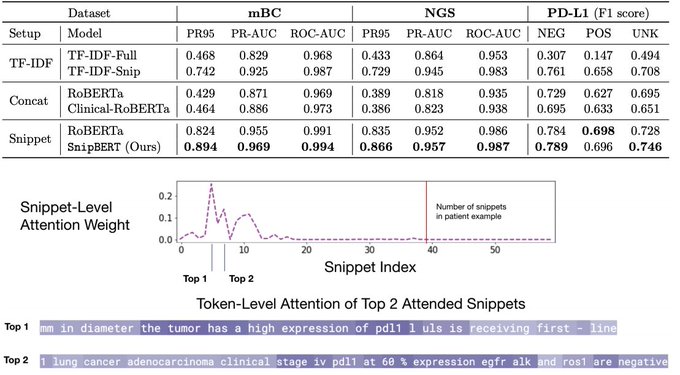
How to extract a clinical variable out of the entire patient’s history notes of > 200K words, using an expensive BERT-style model?
Happy to share SnipBERT done
@flatironhealth
in NeurIPS ML4H!
- up to >20% prediction gain
- clues for model interpretation
Details ⬇️
2
4
19
Excited to co-organize AI for Science workshop again at NeurIPS 2023!
(1/3) I am thrilled to announce that
@AI_for_Science
workshop is back with
#NeurIPS2023
! This year we put together several new programs with a new theme "from theory to practice", including a panel discussion to align the expectation between academia and funding agencies.
3
36
144
0
0
18
New TDC Leaderboard on docking score molecule generation⬇️
We restrict the Oracle calls to simulate realistic wet-lab constraint; realistic eval metrics such as %pass mol filters and
@MoleculeOne
for synthesizablity! => Most SOTA fail!
Happy to announce our latest
#benchmark
and
#leaderboard
on molecular
#docking
to evaluate approaches for structure-based
#drugdiscovery
and
#molecule
generation
0
9
28
0
1
14
TDC preprint is alive in arXiv! ⬇️
Excited to share preprint on Therapeutics Data Commons!
Paper:
Website:
TDC is a unifying framework across the entire range of
#therapeutics
#ML
. Ecosystem of tools, leaderboards & community resr
66 ML-ready datasets
22 ML tasks
1
62
220
0
2
14
Really excited for my first NeurIPS paper! We introduce a general and effective framework for graph meta learning, check it out: . Thanks Marinka for the great guidance!
Thrilled that our lab has 4/4 papers accepted at
#NeurIPS2020
! Not bad for a lab just 5 months old at submission deadline. Congrats to fantastic students and collaborators
@_michellemli
@xiangzhang1015
@KexinHuang5
@IAmSamFin
@Emily_Alsentzer
@Harvard
@HarvardDBMI
@harvard_data
11
7
182
2
1
14
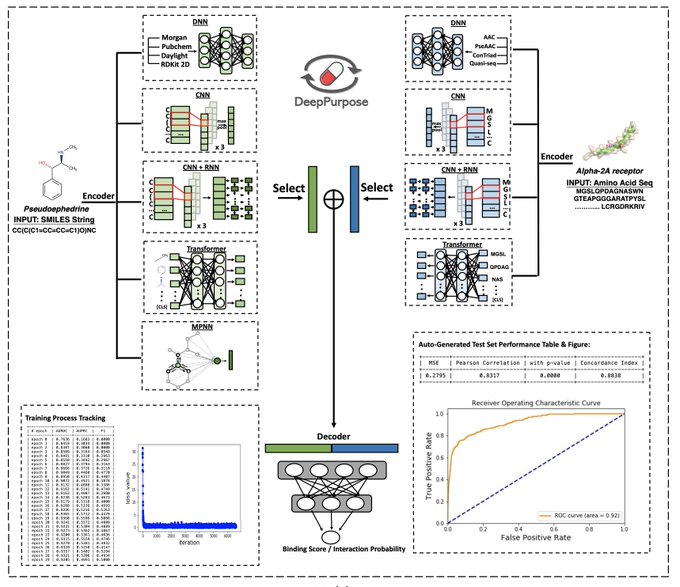
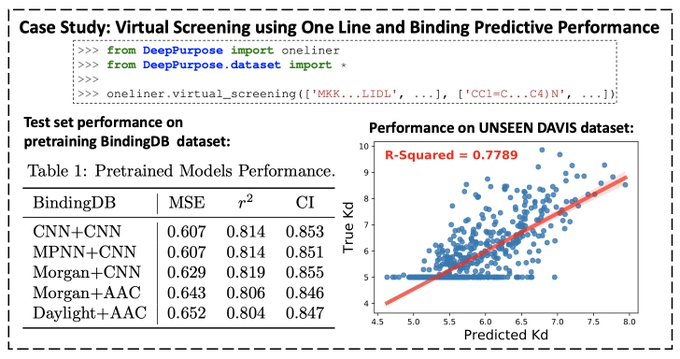
A tutorial of DeepPurpose for data scientists👇
@TianfanFu
@AiLucasg
@marinkazitnik
@caoxiao_danica
@jimeng
@IQVIA_global
@harvard_data
0
2
12
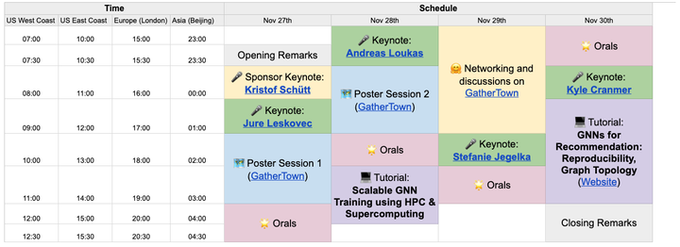
LoG is happening tomorrow! Join now:
LoG is happening tomorrow!
Highlights of the program:
🎤Exciting keynotes from
@jure
,
@andreasloudaros
, Stefanie Jegelka,
@KyleCranmer
,
@ktschuett
🌟 12 orals
💻 Tutorials on scalability & recommendation
🤗 poster sessions & networking
Join now via
1
32
104
0
0
11
Very happy to work with and learnt a ton from
@YingJin531
Emmanuel and
@jure
on this project.
Guaranteed uncertainty has extensive applications in graph ML, biomedical ML, drug discovery ML, that’s why I am excited about it! Stay tuned for our future work in this direction!
1
1
8
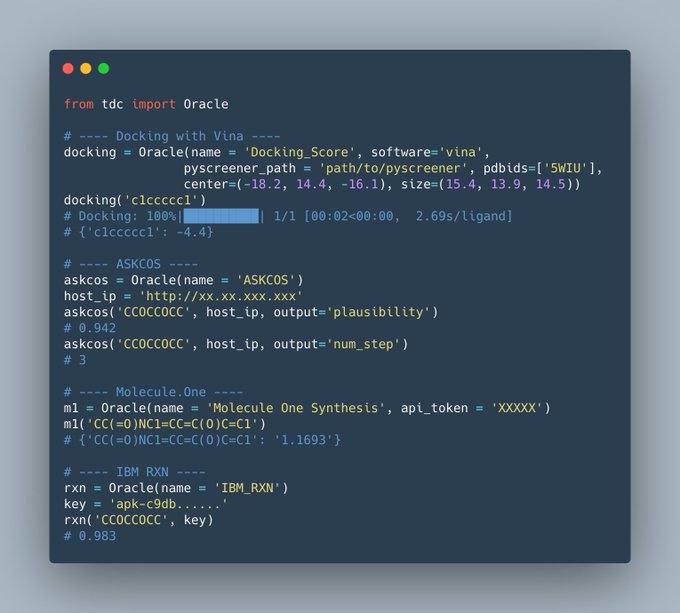
We just released four new realistic oracles on docking scores and synthetic pathway analysis from ASKCOS,
@MoleculeOne
, and
@ForRxn
, all under one-line-of-code!
“The current evaluations for generative models do not reflect the complexity of real discovery problems.”
TDC now includes one-liners for four more realistic oracles: docking scores, synthetic accessibility from ASKCOS,
@MoleculeOne
,
@ForRxn
.
Info:
0
7
23
0
2
8
Super happy to see this preprint out! Lucky to work with and learned a ton from amazing collaborators
@marinkazitnik
@payal_chandak
@WangQianwenToo
Shreyas, Akhil,
@jure
@girish_nadkarni
@ngehlenborg
7/7
0
0
7
Joint work with an all-star team
@marinkazitnik
@TianfanFu
@WenhaoGao1
@yzhao062
!
TDC is a community effort and we are looking for contributors! If interested, fill in this form:
Github:
Website:
1
3
7
With an awesome team:
@TianfanFu
@caoxiao_danica
@jimeng
@marinkazitnik
@AiLucasg
!
Btw, a link to TDC if you want to try out the ADME datasets:
0
1
6
Had fun with these amazing folks
@LogmlSchool
where we studied if classic network medicine principle (local hypothesis, shared component hypothesis) is automatically encoded in the bio KG embedding! ⬇️
I am excited to share our
@LogmlSchool
Project summary: Exploring network medicine principles encoded by knowledge graphs embeddings.
Thanks to
@KexinHuang5
@KumailAlhamoud
@farhan__tanvir
Aarthi Venkat and Yepeng Huang for the fun time together!
4
11
39
0
2
6
With
@jimeng
@genomestake
@caoxiao_danica
@AiLucasg
Cathy Critchlow
Github repo that hosts links to all papers following the organization in the paper:
0
4
6
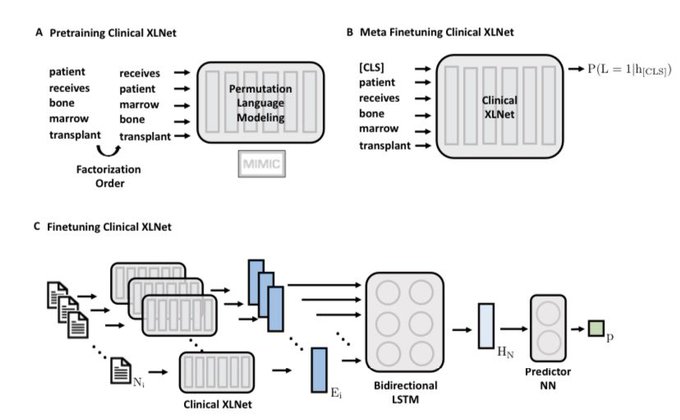
Checkout Clinical XLNet () !
We adapt XLNet to clinical context and leverage the sequential dimension of notes.
We did thorough cohort curation with our awesome clinician team
@DanaFarber
and achieve SOTA on Prolonged Mechanical Ventilation prediction!
1
1
6
Come to poster
#60
on Friday at 5:15PM EST
#ML4H
! Paper at
Enjoyable summer work done at
@flatironhealth
, with the amazing
@sankeerthsai23
and
@alex_s_rich
! This work is built upon many great ideas from the ML team
@benbirnbaum
@GriffinAdams16
,….!
0
2
5
Happy to present SkipGNN! We inject skip-similarity into GNN, which is a very powerful property that distincts molecular networks from classic networks! The result is promising on DDI, PPI, GDI, DTI!
Thanks for all the guidance!
@marinkazitnik
@caoxiao_danica
@jimeng
@AiLucasg
Excited to share a new preprint on predicting molecular
#interactions
with skip-graph networks
Skip-
#GNN
uses 2nd-order interactions, which proved incredibly useful in
#bionets
over the last decade
Led by a fantastic student
@KexinHuang5
@harvard_data
0
7
49
0
1
5
ClinicalBERT is out in () Good performance on intrinsic eval and readmission prediction!
Code and checkpoints:
Greatest logo design by
@thejaan
!
Also, checkout concurrent work by
@Emily_Alsentzer
() !
0
0
4
Great experience as an intern last summer at Pfizer's ML group, learned a lot from
@MBordyuh
@VishnuSresht
Brajesh Rai
0
1
2
Our work on predicting drug interaction with substructure representation is featured in
@techreview
!
0
0
3
We open source all the scripts and weights! Including pre-training at . With
@tremblerz
Sitong Chen
@lindvalllab
et al. from the great HST 953 class
@mit_hst
@MITcriticaldata
0
1
3
Come to our workshop tomorrow! Lots of great talks!
Can't wait to see you at 8am-6pm (EST), Monday! Join us in this great event with 7 featured LIVE talks, a panel discussion and 52 contributed papers about
#AI4Science
@NeurIPSConf
Schedule:
Virtual site:
2
17
83
0
0
2
@andrewwhite01
@marinkazitnik
@WangQianwenToo
@payal_chandak
@ngehlenborg
@icmlconf
@HarvardDBMI
here you go:
0
0
3
@A_Aspuru_Guzik
@curiouswavefn
@neurobongo
SELFIES could also mitigate one weakness of the paper (some decomposed substrings of SMILES are not chemically valid, which may sometimes hurt the explainability). Thanks and will give it a try!
0
0
2
@CyrusMaher
@GradioML
Hey Cyrus, glad to hear it is useful! I contacted the Gradio team to get access, I will PM you the contact person’s email address
1
0
2
@david_sontag
@_MiguelHernan
China currently implements a “COVID QR-code” which stratifies each person’s health status into green, yellow, red, based on self-reported info. The lockdown is lifted only on people with green code. Could it serve as a preliminary example?
0
0
2
@CyrusMaher
Hey Cyrus, we plan to release the project in next Tuesday in will also keep you posted here!
0
0
1
@CyrusMaher
@marinkazitnik
@TianfanFu
@WenhaoGao1
@yzhao062
Hey Cyrus, yes, it is because some source datasets contain this license. So in the first release, we just use the most restricted one. But this is a good point, we will try to find license from individual data source so that industry folks can use them. added to the backlog!
0
0
1
@_romain_lopez_
Thank you for making the internship experience awesome and for all the advice!!
0
0
0
@CyrusMaher
Interesting! I tried to use MLP + Morgan as the drug encoder and it is not as good for DTI, but I indeed found it has comparable and sometimes better performance in many molecular property predictions tasks compared to MPNN and transformers etc. I will try the random forest! Thx!
0
0
1