Luca Marconato
@LucaMarconato2
Followers
620
Following
333
Media
13
Statuses
87
Computational scientist at EMBL in the @OliverStegle lab. Deep learning methods and infrastructures for spatial multi-omics data.
Joined December 2019
The @ChanZuckerberg Cell Science Meeting in Chicago just concluded. I had the opportunity to present the latest advancements in spatial omics within the #scverse ecosystem, highlighting our efforts to scale our infrastructure in preparation for the datasets of tomorrow.
0
1
12
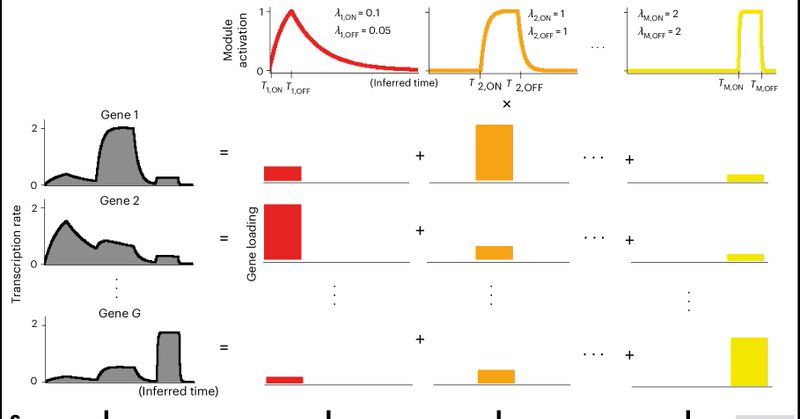
RT @bayraktar_lab: happy to see our cell2fate model out at @naturemethods. It is a completely new take on RNA velocity to disentangle compl….
nature.com
Nature Methods - Cell2fate improves RNA velocity analysis of single-cell and spatial transcriptomics data by module decomposition of realistic biophysical models of transcription dynamics.
0
39
0
RT @ICGEBCapeTown: Join our next #ICGEBCapeTown International seminar by @LucaMarconato2 from @embl @OliverStegle Lab. 📅 25 February 202….
0
5
0
Join us in Paris at the @OwkinScience headquarters to work together on spatial omics! #scverse.
Owkin and @scverse_team are excited to announce their upcoming hackathon, focused on spatial omics data analysis. 📅 March 17-19, 2025.📍 Owkin office, Paris. If you are a bioinformatician interested in spatial biology, this is your chance to make a contribution to open-source
0
3
7
RT @OwkinScience: Owkin and @scverse_team are excited to announce their upcoming hackathon, focused on spatial omics data analysis. 📅 Marc….
0
7
0
The latest #scverse hackathon on spatial omics has really been a blast! A small example: Day 2 was planned from 8:30 am to 6:00 pm, yet we had people hacking until 9-10 pm! (pics 2-3). Looking forward to the new in-person event!.
Recently, we held the first #scverse SpatialData 3-day hackathon in Basel (Switzerland), where we brought together 20 developers from the Python, R, and web communities to work on interoperability, performance, and ergonomics for spatial omics data. Here is what we worked on!
0
2
13
I had the opportunity to present my work at BIOINFO 2024 in Gyeongju, Korea. It was great to meet previous colleagues from Heidelberg, @pjb7687 and @hyobin_jeong, and be welcomed by such a vibrant research community!
@LucaMarconato2 is giving a super nice tutorial on SpatialData @ BIOINFO2024, KSBi in Gyeongju, Korea.
0
0
16
In September, I represented the SpatialData team at SCG 2024. I was honored to receive a best poster award; I want to thank my team (in particular @g_palla1, Tim, @WVierdag, @ky396, Sonja, . ) who made the project possible. In the pic, @gtcaa and @MKoutrouli from @scverse_team.
0
2
25
RT @brianfclarke: 🧵 1/9 Delighted to announce that #DeepRVAT is out today in Nature Genetics! Here's a recap of our tweetorial from the pre….
0
26
0
The recording for today's webinar will soon be available from @EBItraining. Meanwhile here are the notebooks 🤓 Thanks again for inviting me and @WVierdag, and thanks to the @scverse_team for the collaborative effort in preparing the notebook.
github.com
Contribute to PMBio/spatialdata-workshops development by creating an account on GitHub.
There’s still time to join tomorrow’s #webinar about SpatialData, an #OpenAccess data framework for spatial omics. Registration is free but essential: #bioinformatics #datascience #omics #multiomics #transcriptomics #singlecell #spatialomics
0
6
23
RT @QuentinBlampey: Happy to share Novae, our new foundation model for spatial transcriptomics data 💫.Load a pre-trained model and start us….
github.com
Graph-based foundation model for spatial transcriptomics data. Zero-shot spatial domain inference, batch-effect correction, and many other features. - MICS-Lab/novae
0
7
0
RT @MKoutrouli: @scverse_team is at #SCG2024 @LucaMarconato2 @gtcaa .Thanks to the organising committee. The first day was already EXCELLEN….
0
3
0
RT @MKoutrouli: First conference, first selfie with some of the incredible @scverse_team core and volunteers! Feeling so grateful to be par….
0
7
0
RT @WVierdag: Awesome week last week representing both @napari_imaging and #SpatialData @scverse at @EuroSciPy2024. It was great to meet ma….
0
9
0
Which is more convenient: viewing the "Visium HD bins" as a collection of millions of vector geometries, or as a large multi-channel image given their grid arrangement?. In the webinar, @QuentinBlampey, @WVierdag, and I will discuss code and use cases for both approaches.
Your reaction to #VisiumHD data analysis may be somewhere between 🤓 and 😵. Wherever you land, join this webinar to see how intuitive and adaptable high-def #spatialtranscriptomics data exploration can be with tools like Bin2Cell & SpatialData.
0
13
43
RT @joel_luethi: It's great to be at #EMC2024 and learn more about all the different microscopy techniques. I will be presenting a poster o….
0
15
0
RT @bayraktar_lab: WebAtlas for visualising single cell & spatial data online is out: With @Muzz_Haniffa @BioinfoTo….
0
72
0
RT @Severin_1991: 🌟 Join me at #Scverse2024 for the "GPU-Accelerated Single-Cell Analysis with rapids-singlecell" workshop! .📍 Munich, Germ….
github.com
rapids_singlecell: GPU-accelerated framework for scRNA analysis - scverse/rapids_singlecell
0
11
0
RT @scverse_team: 📢 We're thrilled to announce that we are offering travel grants to cover registration fees and travel expenses for our #s….
0
9
0